Degron on:
[Wikipedia]
[Google]
[Amazon]
 A degron is a portion of a protein that is important in regulation of protein degradation rates. Known degrons include short amino acid sequences, structural motifs and exposed amino acids (often Lysine or Arginine) located anywhere in the protein. In fact, some proteins can even contain multiple degrons. Degrons are present in a variety of organisms, from the N-degrons (see
A degron is a portion of a protein that is important in regulation of protein degradation rates. Known degrons include short amino acid sequences, structural motifs and exposed amino acids (often Lysine or Arginine) located anywhere in the protein. In fact, some proteins can even contain multiple degrons. Degrons are present in a variety of organisms, from the N-degrons (see
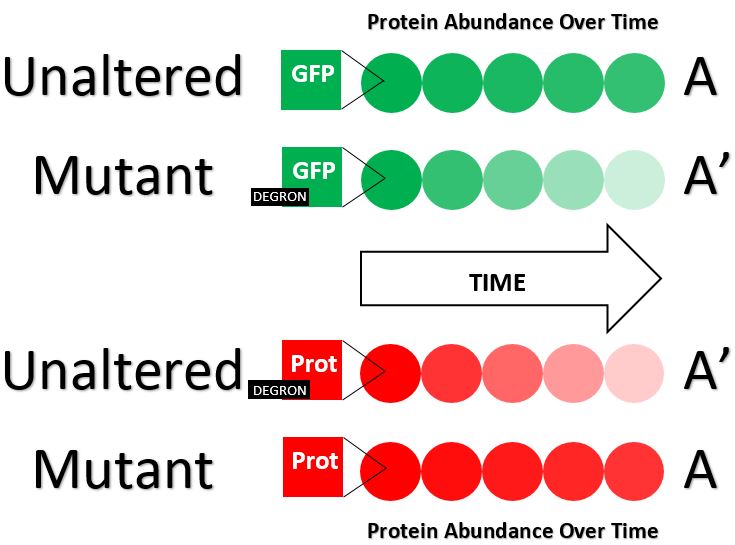 In order to identify a portion of a protein as a degron, there are often three steps performed. First, the degron candidate is fused to a stable protein, such as GFP, and protein abundances over time are compared between the unaltered protein and the fusion (as shown in green). If the candidate is in fact a degron, then the abundance of the fusion protein will decrease much faster than that of the unaltered protein. Second, a mutant form of the degron's protein is designed such that it lacks the degron candidate. Similar to before, the abundance of the mutant protein over time is compared to that of the unaltered protein (as shown in red). If the deleted degron candidate is in fact a degron, then the mutant protein abundance will decrease much slower than that of the unaltered protein. Recall that degrons are often referred to as “Ubiquitin-dependent” or “Ubiquitin-independent” The third step performed is often done after one or both of the previous two steps, because it serves to identify the Ubiquitin dependence or lack thereof of a previously identified degron. In this step, protein A and A’ (identical in every way except the presence of degron in A’) will be examined. Note that mutation or fusion procedures could be performed here, so either A is a protein like GFP and A’ is a fusion of GFP with the degron (as shown in green) or A’ is the degron's protein and A is a mutant form without the degron (as shown in Red.) The amount of Ubiquitin bound to A and to A’ will be measured. A significant increase in the amount of Ubiquitin in A’ as compared to A will suggest that the degron is Ubiquitin-dependent.
In order to identify a portion of a protein as a degron, there are often three steps performed. First, the degron candidate is fused to a stable protein, such as GFP, and protein abundances over time are compared between the unaltered protein and the fusion (as shown in green). If the candidate is in fact a degron, then the abundance of the fusion protein will decrease much faster than that of the unaltered protein. Second, a mutant form of the degron's protein is designed such that it lacks the degron candidate. Similar to before, the abundance of the mutant protein over time is compared to that of the unaltered protein (as shown in red). If the deleted degron candidate is in fact a degron, then the mutant protein abundance will decrease much slower than that of the unaltered protein. Recall that degrons are often referred to as “Ubiquitin-dependent” or “Ubiquitin-independent” The third step performed is often done after one or both of the previous two steps, because it serves to identify the Ubiquitin dependence or lack thereof of a previously identified degron. In this step, protein A and A’ (identical in every way except the presence of degron in A’) will be examined. Note that mutation or fusion procedures could be performed here, so either A is a protein like GFP and A’ is a fusion of GFP with the degron (as shown in green) or A’ is the degron's protein and A is a mutant form without the degron (as shown in Red.) The amount of Ubiquitin bound to A and to A’ will be measured. A significant increase in the amount of Ubiquitin in A’ as compared to A will suggest that the degron is Ubiquitin-dependent.
 A degron is a portion of a protein that is important in regulation of protein degradation rates. Known degrons include short amino acid sequences, structural motifs and exposed amino acids (often Lysine or Arginine) located anywhere in the protein. In fact, some proteins can even contain multiple degrons. Degrons are present in a variety of organisms, from the N-degrons (see
A degron is a portion of a protein that is important in regulation of protein degradation rates. Known degrons include short amino acid sequences, structural motifs and exposed amino acids (often Lysine or Arginine) located anywhere in the protein. In fact, some proteins can even contain multiple degrons. Degrons are present in a variety of organisms, from the N-degrons (see N-end Rule The ''N''-end rule is a rule that governs the rate of protein degradation through recognition of the N-terminal residue of proteins. The rule states that the ''N''-terminal amino acid of a protein determines its half-life (time after which half of ...
) first characterized in yeast to the PEST sequence
A PEST sequence is a peptide sequence that is rich in proline (P), glutamic acid (E), serine (S), and threonine (T). This sequence is associated with proteins that have a short intracellular half-life; therefore, it is hypothesized that the PEST se ...
of mouse ornithine decarboxylase. Degrons have been identified in prokaryote
A prokaryote () is a single-celled organism that lacks a nucleus and other membrane-bound organelles. The word ''prokaryote'' comes from the Greek πρό (, 'before') and κάρυον (, 'nut' or 'kernel').Campbell, N. "Biology:Concepts & Connec ...
s as well as eukaryotes
Eukaryotes () are organisms whose cells have a nucleus. All animals, plants, fungi, and many unicellular organisms, are Eukaryotes. They belong to the group of organisms Eukaryota or Eukarya, which is one of the three domains of life. Bacte ...
. While there are many types of different degrons, and a high degree of variability even within these groups, degrons are all similar for their involvement in regulating the rate of a protein's degradation. Much like protein degradation (see proteolysis
Proteolysis is the breakdown of proteins into smaller polypeptides or amino acids. Uncatalysed, the hydrolysis of peptide bonds is extremely slow, taking hundreds of years. Proteolysis is typically catalysed by cellular enzymes called protease ...
) mechanisms are categorized by their dependence or lack thereof on Ubiquitin
Ubiquitin is a small (8.6 kDa) regulatory protein found in most tissues of eukaryotic organisms, i.e., it is found ''ubiquitously''. It was discovered in 1975 by Gideon Goldstein and further characterized throughout the late 1970s and 1980s. Fo ...
, a small protein involved in proteasomal protein degradation, Degrons may also be referred to as “Ubiquitin-dependent" or “Ubiquitin-independent".
Types
Ubiquitin-dependent degrons are so named because they are implicated in the polyubiquitination process for targeting a protein to the proteasome. In some cases, the degron itself serves as the site for polyubiquitination as is seen inTAZ Taz or TAZ may refer to:
Geography
*Taz (river), a river in western Siberia, Russia
*Taz Estuary, the estuary of the river Taz in Russia
People
* Taz people, an ethnic group in Russia
** Taz language, a form of Northeastern Mandarin spoken by ...
and β-catenin
Catenin beta-1, also known as beta-catenin (β-catenin), is a protein that in humans is encoded by the ''CTNNB1'' gene.
Beta-catenin is a dual function protein, involved in regulation and coordination of cell–cell adhesion and gene transcriptio ...
proteins. Because the exact mechanism by which a degron is involved in a protein's polyubiqutination is not always known, degrons are classified as Ubiquitin-dependent if their removal from the protein leads to less ubiquitination or if their addition to another protein leads to more ubiquitination.
In contrast, Ubiquitin-independent degrons are not necessary for the polyubiquitination of their protein. For example, the degron on IkBa, a protein involved in the regulation of the immune system, was not shown to be involved in ubiquitination since its addition to Green Fluorescent Protein (GFP GFP may refer to:
Organisations
* Gaelic Football Provence, a French Gaelic Athletic Association club
* Geheime Feldpolizei, the German secret military police during the Second World War
* French Group for the Study of Polymers and their Applicat ...
) did not increase ubiquitination. However, a degron can only hint at the mechanism by which a protein is degraded and so identifying and classifying a degron is only the first step in understanding the degradation process for its protein.
Identification
 In order to identify a portion of a protein as a degron, there are often three steps performed. First, the degron candidate is fused to a stable protein, such as GFP, and protein abundances over time are compared between the unaltered protein and the fusion (as shown in green). If the candidate is in fact a degron, then the abundance of the fusion protein will decrease much faster than that of the unaltered protein. Second, a mutant form of the degron's protein is designed such that it lacks the degron candidate. Similar to before, the abundance of the mutant protein over time is compared to that of the unaltered protein (as shown in red). If the deleted degron candidate is in fact a degron, then the mutant protein abundance will decrease much slower than that of the unaltered protein. Recall that degrons are often referred to as “Ubiquitin-dependent” or “Ubiquitin-independent” The third step performed is often done after one or both of the previous two steps, because it serves to identify the Ubiquitin dependence or lack thereof of a previously identified degron. In this step, protein A and A’ (identical in every way except the presence of degron in A’) will be examined. Note that mutation or fusion procedures could be performed here, so either A is a protein like GFP and A’ is a fusion of GFP with the degron (as shown in green) or A’ is the degron's protein and A is a mutant form without the degron (as shown in Red.) The amount of Ubiquitin bound to A and to A’ will be measured. A significant increase in the amount of Ubiquitin in A’ as compared to A will suggest that the degron is Ubiquitin-dependent.
In order to identify a portion of a protein as a degron, there are often three steps performed. First, the degron candidate is fused to a stable protein, such as GFP, and protein abundances over time are compared between the unaltered protein and the fusion (as shown in green). If the candidate is in fact a degron, then the abundance of the fusion protein will decrease much faster than that of the unaltered protein. Second, a mutant form of the degron's protein is designed such that it lacks the degron candidate. Similar to before, the abundance of the mutant protein over time is compared to that of the unaltered protein (as shown in red). If the deleted degron candidate is in fact a degron, then the mutant protein abundance will decrease much slower than that of the unaltered protein. Recall that degrons are often referred to as “Ubiquitin-dependent” or “Ubiquitin-independent” The third step performed is often done after one or both of the previous two steps, because it serves to identify the Ubiquitin dependence or lack thereof of a previously identified degron. In this step, protein A and A’ (identical in every way except the presence of degron in A’) will be examined. Note that mutation or fusion procedures could be performed here, so either A is a protein like GFP and A’ is a fusion of GFP with the degron (as shown in green) or A’ is the degron's protein and A is a mutant form without the degron (as shown in Red.) The amount of Ubiquitin bound to A and to A’ will be measured. A significant increase in the amount of Ubiquitin in A’ as compared to A will suggest that the degron is Ubiquitin-dependent.
References
{{reflistSee also
*N-end rule The ''N''-end rule is a rule that governs the rate of protein degradation through recognition of the N-terminal residue of proteins. The rule states that the ''N''-terminal amino acid of a protein determines its half-life (time after which half of ...
* Proteasome
Proteasomes are protein complexes which degrade unneeded or damaged proteins by proteolysis, a chemical reaction that breaks peptide bonds. Enzymes that help such reactions are called proteases.
Proteasomes are part of a major mechanism by w ...
* proteolysis
Proteolysis is the breakdown of proteins into smaller polypeptides or amino acids. Uncatalysed, the hydrolysis of peptide bonds is extremely slow, taking hundreds of years. Proteolysis is typically catalysed by cellular enzymes called protease ...
Peptide sequences