CRISPR on:
[Wikipedia]
[Google]
[Amazon]
CRISPR () (an acronym for clustered regularly interspaced short palindromic repeats) is a family of DNA sequences found in the  Cas9 (or "CRISPR-associated protein 9") is an
Cas9 (or "CRISPR-associated protein 9") is an
 In 2005, three independent research groups showed that some CRISPR spacers are derived from phage DNA and
In 2005, three independent research groups showed that some CRISPR spacers are derived from phage DNA and
 The CRISPR-Cas9 system has shown to make effective gene edits in Human tripronuclear zygotes first described in a 2015 paper by Chinese scientists P. Liang and Y. Xu. The system made a successful cleavage of mutant Beta-Hemoglobin (HBB) in 28 out of 54 embryos. 4 out of the 28 embryos were successfully recombined using a donor template given by the scientists. The scientists showed that during DNA recombination of the cleaved strand, the homologous endogenous sequence HBD competes with the exogenous donor template. DNA repair in human embryos is much more complicated and particular than in derived stem cells.
The CRISPR-Cas9 system has shown to make effective gene edits in Human tripronuclear zygotes first described in a 2015 paper by Chinese scientists P. Liang and Y. Xu. The system made a successful cleavage of mutant Beta-Hemoglobin (HBB) in 28 out of 54 embryos. 4 out of the 28 embryos were successfully recombined using a donor template given by the scientists. The scientists showed that during DNA recombination of the cleaved strand, the homologous endogenous sequence HBD competes with the exogenous donor template. DNA repair in human embryos is much more complicated and particular than in derived stem cells.
Image:RF01315.png, ;CRISPR-DR2: Secondary structure taken from th
Rfam
database. Famil
RF01315
Image:RF01318.png, ;CRISPR-DR5: Secondary structure taken from th
Rfam
database. Famil
RF011318
Image:RF01319.png, ;CRISPR-DR6: Secondary structure taken from th
Rfam
database. Famil
RF01319
Image:RF01321.png, ;CRISPR-DR8: Secondary structure taken from th
Rfam
database. Famil
RF01321
Image:RF01322.png, ;CRISPR-DR9: Secondary structure taken from th
Rfam
database. Famil
RF01322
Image:RF01332.png, ;CRISPR-DR19: Secondary structure taken from th
Rfam
database. Famil
RF01332
Image:RF01350.png, ;CRISPR-DR41: Secondary structure taken from th
Rfam
database. Famil
RF01350
Image:RF01365.png, ;CRISPR-DR52: Secondary structure taken from th
Rfam
database. Famil
RF01365
Image:RF01370.png, ;CRISPR-DR57: Secondary structure taken from th
Rfam
database. Famil
RF01370
Image:RF01378.png, ;CRISPR-DR65: Secondary structure taken from th
Rfam
database. Famil
RF01378
 CRISPR-Cas immunity is a natural process of bacteria and archaea. CRISPR-Cas prevents bacteriophage infection,
CRISPR-Cas immunity is a natural process of bacteria and archaea. CRISPR-Cas prevents bacteriophage infection,
 CRISPR associated nucleases have shown to be useful as a tool for molecular testing due to their ability to specifically target nucleic acid sequences in a high background of non-target sequences. In 2016, the Cas9 nuclease was used to deplete unwanted nucleotide sequences in next-generation sequencing libraries while requiring only 250 picograms of initial RNA input. Beginning in 2017, CRISPR associated nucleases were also used for direct diagnostic testing of nucleic acids, down to single molecule sensitivity. CRISPR diversity is used as an analysis target to discern
CRISPR associated nucleases have shown to be useful as a tool for molecular testing due to their ability to specifically target nucleic acid sequences in a high background of non-target sequences. In 2016, the Cas9 nuclease was used to deplete unwanted nucleotide sequences in next-generation sequencing libraries while requiring only 250 picograms of initial RNA input. Beginning in 2017, CRISPR associated nucleases were also used for direct diagnostic testing of nucleic acids, down to single molecule sensitivity. CRISPR diversity is used as an analysis target to discern
genome
In the fields of molecular biology and genetics, a genome is all the genetic information of an organism. It consists of nucleotide sequences of DNA (or RNA in RNA viruses). The nuclear genome includes protein-coding genes and non-coding ...
s of prokaryotic organisms such as bacteria
Bacteria (; singular: bacterium) are ubiquitous, mostly free-living organisms often consisting of one biological cell. They constitute a large domain of prokaryotic microorganisms. Typically a few micrometres in length, bacteria were am ...
and archaea. These sequences are derived from DNA fragments of bacteriophage
A bacteriophage (), also known informally as a ''phage'' (), is a duplodnaviria virus that infects and replicates within bacteria and archaea. The term was derived from "bacteria" and the Greek φαγεῖν ('), meaning "to devour". Bact ...
s that had previously infected the prokaryote. They are used to detect and destroy DNA from similar bacteriophages during subsequent infections. Hence these sequences play a key role in the antiviral (i.e. anti-phage) defense system of prokaryotes and provide a form of acquired immunity. CRISPR is found in approximately 50% of sequenced bacterial genomes and nearly 90% of sequenced archaea.
 Cas9 (or "CRISPR-associated protein 9") is an
Cas9 (or "CRISPR-associated protein 9") is an enzyme
Enzymes () are proteins that act as biological catalysts by accelerating chemical reactions. The molecules upon which enzymes may act are called substrate (chemistry), substrates, and the enzyme converts the substrates into different molecule ...
that uses CRISPR sequences as a guide to recognize and cleave specific strands of DNA that are complementary to the CRISPR sequence. Cas9 enzymes together with CRISPR sequences form the basis of a technology known as CRISPR-Cas9
Cas9 (CRISPR associated protein 9, formerly called Cas5, Csn1, or Csx12) is a 160 kilodalton protein which plays a vital role in the immunological defense of certain bacteria against DNA viruses and plasmids, and is heavily utilized in geneti ...
that can be used to edit genes within organisms. This editing process has a wide variety of applications including basic biological research, development of biotechnological products, and treatment of diseases. The development of the CRISPR-Cas9 genome editing technique was recognized by the Nobel Prize in Chemistry
)
, image = Nobel Prize.png
, alt = A golden medallion with an embossed image of a bearded man facing left in profile. To the left of the man is the text "ALFR•" then "NOBEL", and on the right, the text (smaller) "NAT•" then "M ...
in 2020 which was awarded to Emmanuelle Charpentier and Jennifer Doudna. In 2022, based on evidence presented in Interference 106, 115, the PTAB tipped the scale for invention covering application of CRISPR-Cas9 in eukaryotic cells in favour of Feng Zhang, a professor of the Broad Institute.
History
Repeated sequences
The discovery of clustered DNA repeats took place independently in three parts of the world. The first description of what would later be called CRISPR is from Osaka University researcher Yoshizumi Ishino and his colleagues in 1987. They accidentally cloned part of a CRISPR sequence together with the "''iap" gene'' ''(isozyme conversion of alkaline phosphatase)'' from the genome of ''Escherichia coli
''Escherichia coli'' (),Wells, J. C. (2000) Longman Pronunciation Dictionary. Harlow ngland Pearson Education Ltd. also known as ''E. coli'' (), is a Gram-negative, facultative anaerobic, rod-shaped, coliform bacterium of the genus '' Esc ...
'' which was their target. The organization of the repeats was unusual. Repeated sequences are typically arranged consecutively, without interspersing different sequences. They did not know the function of the interrupted clustered repeats.
In 1993, researchers of '' Mycobacterium tuberculosis'' in the Netherlands published two articles about a cluster of interrupted direct repeats (DR) in that bacterium. They recognized the diversity of the sequences that intervened in the direct repeats among different strains of ''M. tuberculosis'' and used this property to design a typing method that was named '' spoligotyping'', which is still in use, today.
Francisco Mojica at the University of Alicante in Spain studied repeats observed in the archaeal organisms of '' Haloferax'' and '' Haloarcula'' species and their function. Mojica's supervisor surmised at the time that the clustered repeats had a role in correctly segregating replicated DNA into daughter cells during cell division because plasmids and chromosomes with identical repeat arrays could not coexist in '' Haloferax volcanii''. Transcription of the interrupted repeats was also noted for the first time; this was the first full characterization of CRISPR. By 2000, Mojica performed a survey of scientific literature and one of his students performed a search in published genomes with a program devised by himself. They identified interrupted repeats in 20 species of microbes as belonging to the same family. Because those sequences were interspaced, Mojica initially called these sequences "short regularly spaced repeats" (SRSR). In 2001, Mojica and Ruud Jansen, who were searching for additional interrupted repeats, proposed the acronym CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) to alleviate the confusion stemming from the numerous acronyms used to describe the sequences in the scientific literature. In 2002, Tang, et al. showed evidence that CRISPR repeat regions from the genome of '' Archaeoglobus fulgidus'' were transcribed into long RNA molecules that were subsequently processed into unit-length small RNAs, plus some longer forms of 2, 3, or more spacer-repeat units.
In 2005, yogurt
Yogurt (; , from tr, yoğurt, also spelled yoghurt, yogourt or yoghourt) is a food produced by bacterial fermentation of milk. The bacteria used to make yogurt are known as ''yogurt cultures''. Fermentation of sugars in the milk by these bact ...
researcher Rodolphe Barrangou discovered that '' Streptococcus thermophilus'', after iterative phage challenges, develops increased phage resistance, and this enhanced resistance is due to the incorporation of additional CRISPR spacer sequences. The Danish food company Danisco, which at that time Barrangou worked for, then developed phage-resistant ''S. thermophilus'' strains for use in yogurt production. Danisco was later bought out by DuPont, which "owns about 50 percent of the global dairy culture market" and the technology went mainstream.
CRISPR-associated systems
A major addition to the understanding of CRISPR came with Jansen's observation that the prokaryote repeat cluster was accompanied by a set of homologous genes that make up CRISPR-associated systems or ''cas'' genes. Four ''cas'' genes (''cas'' 1–4) were initially recognized. The Cas proteins showedhelicase
Helicases are a class of enzymes thought to be vital to all organisms. Their main function is to unpack an organism's genetic material. Helicases are motor proteins that move directionally along a nucleic acid phosphodiester backbone, separat ...
and nuclease motifs, suggesting a role in the dynamic structure of the CRISPR loci. In this publication, the acronym CRISPR was used as the universal name of this pattern. However, the CRISPR function remained enigmatic.
 In 2005, three independent research groups showed that some CRISPR spacers are derived from phage DNA and
In 2005, three independent research groups showed that some CRISPR spacers are derived from phage DNA and extrachromosomal DNA
Extrachromosomal DNA (abbreviated ecDNA) is any DNA that is found off the chromosomes, either inside or outside the nucleus of a cell. Most DNA in an individual genome is found in chromosomes contained in the nucleus. Multiple forms of extrachromo ...
such as plasmids. In effect, the spacers are fragments of DNA gathered from viruses that previously tried to attack the cell. The source of the spacers was a sign that the CRISPR/''cas'' system could have a role in adaptive immunity in bacteria
Bacteria (; singular: bacterium) are ubiquitous, mostly free-living organisms often consisting of one biological cell. They constitute a large domain of prokaryotic microorganisms. Typically a few micrometres in length, bacteria were am ...
. All three studies proposing this idea were initially rejected by high-profile journals, but eventually appeared in other journals.
The first publication proposing a role of CRISPR-Cas in microbial immunity, by Mojica and collaborators at the University of Alicante, predicted a role for the RNA transcript of spacers on target recognition in a mechanism that could be analogous to the RNA interference system used by eukaryotic cells. Koonin and colleagues extended this RNA interference hypothesis by proposing mechanisms of action for the different CRISPR-Cas subtypes according to the predicted function of their proteins.
Experimental work by several groups revealed the basic mechanisms of CRISPR-Cas immunity. In 2007, the first experimental evidence that CRISPR was an adaptive immune system was published. A CRISPR region in '' Streptococcus thermophilus'' acquired spacers from the DNA of an infecting bacteriophage
A bacteriophage (), also known informally as a ''phage'' (), is a duplodnaviria virus that infects and replicates within bacteria and archaea. The term was derived from "bacteria" and the Greek φαγεῖν ('), meaning "to devour". Bact ...
. The researchers manipulated the resistance of ''S. thermophilus'' to different types of phages by adding and deleting spacers whose sequence matched those found in the tested phages. In 2008, Brouns and Van der Oost identified a complex of Cas proteins (called Cascade) that in ''E. coli'' cut the CRISPR RNA precursor within the repeats into mature spacer-containing RNA molecules called CRISPR RNA
CRISPR RNA or crRNA is a RNA transcript from the CRISPR locus. CRISPR-Cas (clustered, regularly interspaced short palindromic repeats - CRISPR associated systems) is an adaptive immune system found in bacteria and archaea to protect against mobil ...
(crRNA), which remained bound to the protein complex. Moreover, it was found that Cascade, crRNA and a helicase/nuclease ( Cas3) were required to provide a bacterial host with immunity against infection by a DNA virus. By designing an anti-virus CRISPR, they demonstrated that two orientations of the crRNA (sense/antisense) provided immunity, indicating that the crRNA guides were targeting dsDNA. That year Marraffini and Sontheimer confirmed that a CRISPR sequence of ''S. epidermidis
''Staphylococcus epidermidis'' is a Gram-positive bacterium, and one of over 40 species belonging to the genus ''Staphylococcus''. It is part of the normal human microbiota, typically the skin microbiota, and less commonly the mucosal microbiota ...
'' targeted DNA and not RNA to prevent conjugation
Conjugation or conjugate may refer to:
Linguistics
*Grammatical conjugation, the modification of a verb from its basic form
* Emotive conjugation or Russell's conjugation, the use of loaded language
Mathematics
*Complex conjugation, the change ...
. This finding was at odds with the proposed RNA-interference-like mechanism of CRISPR-Cas immunity, although a CRISPR-Cas system that targets foreign RNA was later found in '' Pyrococcus furiosus''. A 2010 study showed that CRISPR-Cas cuts both strands of phage and plasmid DNA in ''S. thermophilus''.
Cas9
A simpler CRISPR system from ''Streptococcus pyogenes
''Streptococcus pyogenes'' is a species of Gram-positive, aerotolerant bacteria in the genus '' Streptococcus''. These bacteria are extracellular, and made up of non-motile and non-sporing cocci (round cells) that tend to link in chains. They ...
'' relies on the protein Cas9. The Cas9 endonuclease
Endonucleases are enzymes that cleave the phosphodiester bond within a polynucleotide chain. Some, such as deoxyribonuclease I, cut DNA relatively nonspecifically (without regard to sequence), while many, typically called restriction endonuclease ...
is a four-component system that includes two small molecules: crRNA and trans-activating CRISPR RNA (tracrRNA). Jennifer Doudna and Emmanuelle Charpentier re-engineered the Cas9 endonuclease into a more manageable two-component system by fusing the two RNA molecules into a "single-guide RNA" that, when combined with Cas9, could find and cut the DNA target specified by the guide RNA. This contribution was so significant that it was recognized by the Nobel Prize in Chemistry
)
, image = Nobel Prize.png
, alt = A golden medallion with an embossed image of a bearded man facing left in profile. To the left of the man is the text "ALFR•" then "NOBEL", and on the right, the text (smaller) "NAT•" then "M ...
in 2020. By manipulating the nucleotide sequence of the guide RNA, the artificial Cas9 system could be programmed to target any DNA sequence for cleavage. Another group of collaborators comprising Virginijus Šikšnys together with Gasiūnas, Barrangou, and Horvath showed that Cas9 from the ''S. thermophilus'' CRISPR system can also be reprogrammed to target a site of their choosing by changing the sequence of its crRNA. These advances fueled efforts to edit genomes with the modified CRISPR-Cas9 system.
Groups led by Feng Zhang and George Church simultaneously published descriptions of genome editing in human cell cultures using CRISPR-Cas9 for the first time. It has since been used in a wide range of organisms, including baker's yeast (''Saccharomyces cerevisiae
''Saccharomyces cerevisiae'' () (brewer's yeast or baker's yeast) is a species of yeast (single-celled fungus microorganisms). The species has been instrumental in winemaking, baking, and brewing since ancient times. It is believed to have been o ...
''), the opportunistic pathogen '' Candida albicans'', zebrafish ('' Danio rerio''), fruit flies (''Drosophila melanogaster
''Drosophila melanogaster'' is a species of fly (the taxonomic order Diptera) in the family Drosophilidae. The species is often referred to as the fruit fly or lesser fruit fly, or less commonly the " vinegar fly" or " pomace fly". Starting with ...
''), ants ('' Harpegnathos saltator'' and '' Ooceraea biroi''), mosquitoes ('' Aedes aegypti''), nematodes (''Caenorhabditis elegans
''Caenorhabditis elegans'' () is a free-living transparent nematode about 1 mm in length that lives in temperate soil environments. It is the type species of its genus. The name is a blend of the Greek ''caeno-'' (recent), ''rhabditis'' (r ...
''), plants, mice ('' Mus musculus domesticus)'', monkeys and human embryos.
CRISPR has been modified to make programmable transcription factors that allows targeting and activation or silencing specific genes.
Cas12a
In 2015, the nuclease Cas12a (formerly known as ) was characterized in the CRISPR/Cpf1 system of the bacterium '' Francisella novicida''. Its original name, from a TIGRFAMs protein family definition built in 2012, reflects the prevalence of its CRISPR-Cas subtype in the ''Prevotella'' and ''Francisella'' lineages. Cas12a showed several key differences from Cas9 including: causing a 'staggered' cut in double stranded DNA as opposed to the 'blunt' cut produced by Cas9, relying on a 'T rich' PAM (providing alternative targeting sites to Cas9), and requiring only a CRISPR RNA (crRNA) for successful targeting. By contrast, Cas9 requires both crRNA and a transactivating crRNA (tracrRNA). These differences may give Cas12a some advantages over Cas9. For example, Cas12a's small crRNAs are ideal for multiplexed genome editing, as more of them can be packaged in one vector than can Cas9's sgRNAs. As well, the sticky 5′ overhangs left by Cas12a can be used for DNA assembly that is much more target-specific than traditional Restriction Enzyme cloning. Finally, Cas12a cleaves DNA 18–23 base pairs downstream from the PAM site. This means there is no disruption to the recognition sequence after repair, and so Cas12a enables multiple rounds of DNA cleavage. By contrast, since Cas9 cuts only 3 base pairs upstream of the PAM site, the NHEJ pathway results in indel mutations that destroy the recognition sequence, thereby preventing further rounds of cutting. In theory, repeated rounds of DNA cleavage should cause an increased opportunity for the desired genomic editing to occur. A distinctive feature of Cas12a, as compared to Cas9, is that after cutting its target, Cas12a remains bound to the target and then cleaves other ssDNA molecules non-discriminately. This property is called "collateral cleavage" or "trans-cleavage" activity and has been exploited for the development of various diagnostic technologies.Cas13
In 2016, the nuclease (formerly known as ) from the bacterium ''Leptotrichia shahii'' was characterized. Cas13 is an RNA-guided RNA endonuclease, which means that it does not cleave DNA, but only single-stranded RNA. Cas13 is guided by its crRNA to a ssRNA target and binds and cleaves the target. Similar to Cas12a, the Cas13 remains bound to the target and then cleaves other ssRNA molecules non-discriminately. This collateral cleavage property has been exploited for the development of various diagnostic technologies. In 2021, Dr. Hui Yang characterized novel miniature Cas13 protein (mCas13) variants, Cas13X & Cas13Y. Using a small portion of N gene sequence from SARS-CoV-2 as a target in characterization of mCas13, revealed the sensitivity and specificity of mCas13 coupled with RT-LAMP for detection of SARS-CoV-2 in both synthetic and clinical samples over other available standard tests like RT-qPCR (1 copy/μL).Locus structure
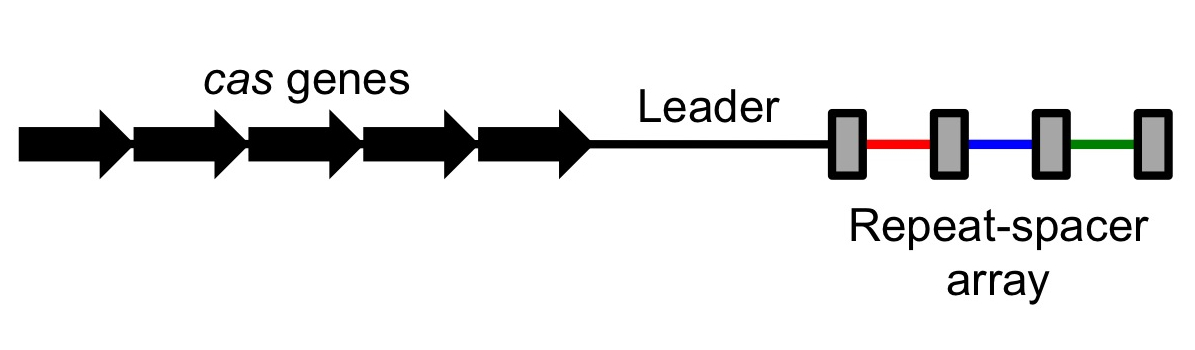
Repeats and spacers
The CRISPR array is made up of an AT-rich leader sequence followed by short repeats that are separated by unique spacers. CRISPR repeats typically range in size from 28 to 37base pairs
A base pair (bp) is a fundamental unit of double-stranded nucleic acids consisting of two nucleobases bound to each other by hydrogen bonds. They form the building blocks of the DNA double helix and contribute to the folded structure of both D ...
(bps), though there can be as few as 23 bp and as many as 55 bp. Some show dyad symmetry, implying the formation of a secondary structure
Protein secondary structure is the three dimensional form of ''local segments'' of proteins. The two most common secondary structural elements are alpha helices and beta sheets, though beta turns and omega loops occur as well. Secondary struct ...
such as a stem-loop
Stem-loop intramolecular base pairing is a pattern that can occur in single-stranded RNA. The structure is also known as a hairpin or hairpin loop. It occurs when two regions of the same strand, usually complementary in nucleotide sequence wh ...
('hairpin') in the RNA, while others are designed to be unstructured. The size of spacers in different CRISPR arrays is typically 32 to 38 bp (range 21 to 72 bp). New spacers can appear rapidly as part of the immune response to phage infection. There are usually fewer than 50 units of the repeat-spacer sequence in a CRISPR array.
CRISPR RNA structures
Rfam
database. Famil
RF01315
Image:RF01318.png, ;CRISPR-DR5: Secondary structure taken from th
Rfam
database. Famil
RF011318
Image:RF01319.png, ;CRISPR-DR6: Secondary structure taken from th
Rfam
database. Famil
RF01319
Image:RF01321.png, ;CRISPR-DR8: Secondary structure taken from th
Rfam
database. Famil
RF01321
Image:RF01322.png, ;CRISPR-DR9: Secondary structure taken from th
Rfam
database. Famil
RF01322
Image:RF01332.png, ;CRISPR-DR19: Secondary structure taken from th
Rfam
database. Famil
RF01332
Image:RF01350.png, ;CRISPR-DR41: Secondary structure taken from th
Rfam
database. Famil
RF01350
Image:RF01365.png, ;CRISPR-DR52: Secondary structure taken from th
Rfam
database. Famil
RF01365
Image:RF01370.png, ;CRISPR-DR57: Secondary structure taken from th
Rfam
database. Famil
RF01370
Image:RF01378.png, ;CRISPR-DR65: Secondary structure taken from th
Rfam
database. Famil
RF01378
Cas genes and CRISPR subtypes
Small clusters of ''cas'' genes are often located next to CRISPR repeat-spacer arrays. Collectively the 93 ''cas'' genes are grouped into 35 families based on sequence similarity of the encoded proteins. 11 of the 35 families form the ''cas'' core, which includes the protein families Cas1 through Cas9. A complete CRISPR-Cas locus has at least one gene belonging to the ''cas'' core. CRISPR-Cas systems fall into two classes. Class 1 systems use a complex of multiple Cas proteins to degrade foreign nucleic acids. Class 2 systems use a single large Cas protein for the same purpose. Class 1 is divided into types I, III, and IV; class 2 is divided into types II, V, and VI. The 6 system types are divided into 19 subtypes. Each type and most subtypes are characterized by a "signature gene" found almost exclusively in the category. Classification is also based on the complement of ''cas'' genes that are present. Most CRISPR-Cas systems have a Cas1 protein. Thephylogeny
A phylogenetic tree (also phylogeny or evolutionary tree Felsenstein J. (2004). ''Inferring Phylogenies'' Sinauer Associates: Sunderland, MA.) is a branching diagram or a tree showing the evolutionary relationships among various biological spe ...
of Cas1 proteins generally agrees with the classification system, but exceptions exist due to module shuffling. Many organisms contain multiple CRISPR-Cas systems suggesting that they are compatible and may share components. The sporadic distribution of the CRISPR/Cas subtypes suggests that the CRISPR/Cas system is subject to horizontal gene transfer during microbial evolution
Evolution is change in the heritable characteristics of biological populations over successive generations. These characteristics are the expressions of genes, which are passed on from parent to offspring during reproduction. Variation ...
.
Mechanism
conjugation
Conjugation or conjugate may refer to:
Linguistics
*Grammatical conjugation, the modification of a verb from its basic form
* Emotive conjugation or Russell's conjugation, the use of loaded language
Mathematics
*Complex conjugation, the change ...
and natural transformation
In category theory, a branch of mathematics, a natural transformation provides a way of transforming one functor into another while respecting the internal structure (i.e., the composition of morphisms) of the categories involved. Hence, a na ...
by degrading foreign nucleic acids that enter the cell.
Spacer acquisition
When a microbe is invaded by abacteriophage
A bacteriophage (), also known informally as a ''phage'' (), is a duplodnaviria virus that infects and replicates within bacteria and archaea. The term was derived from "bacteria" and the Greek φαγεῖν ('), meaning "to devour". Bact ...
, the first stage of the immune response is to capture phage DNA and insert it into a CRISPR locus in the form of a spacer. Cas1 and Cas2 are found in both types of CRISPR-Cas immune systems, which indicates that they are involved in spacer acquisition. Mutation studies confirmed this hypothesis, showing that removal of cas1 or cas2 stopped spacer acquisition, without affecting CRISPR immune response.
Multiple Cas1 proteins have been characterised and their structures resolved. Cas1 proteins have diverse amino acid
Amino acids are organic compounds that contain both amino and carboxylic acid functional groups. Although hundreds of amino acids exist in nature, by far the most important are the alpha-amino acids, which comprise proteins. Only 22 alpha ...
sequences. However, their crystal structures are similar and all purified Cas1 proteins are metal-dependent nucleases/ integrases that bind to DNA in a sequence-independent manner. Representative Cas2 proteins have been characterised and possess either (single strand) ssRNA- or (double strand) dsDNA- specific endoribonuclease activity.
In the I-E system of ''E. coli'' Cas1 and Cas2 form a complex where a Cas2 dimer bridges two Cas1 dimers. In this complex Cas2 performs a non-enzymatic scaffolding role, binding double-stranded fragments of invading DNA, while Cas1 binds the single-stranded flanks of the DNA and catalyses their integration into CRISPR arrays. New spacers are usually added at the beginning of the CRISPR next to the leader sequence creating a chronological record of viral infections. In ''E. coli'' a histone like protein called integration host factor ( IHF), which binds to the leader sequence, is responsible for the accuracy of this integration. IHF also enhances integration efficiency in the type I-F system of ''Pectobacterium atrosepticum
''Pectobacterium atrosepticum'' is a species of bacterium. It is a plant pathogen causing blackleg of potato. Its type strain is CFBP 1526T (=LMG 2386T =NCPPB 549TICMP 1526T. Its genome has been sequenced.
References
Further reading
* Liu ...
''. but in other systems, different host factors may be required
Protospacer adjacent motifs (PAM)
Bioinformatic analysis of regions of phage genomes that were excised as spacers (termed protospacers) revealed that they were not randomly selected but instead were found adjacent to short (3–5 bp) DNA sequences termed protospacer adjacent motifs (PAM). Analysis of CRISPR-Cas systems showed PAMs to be important for type I and type II, but not type III systems during acquisition. In type I and type II systems, protospacers are excised at positions adjacent to a PAM sequence, with the other end of the spacer cut using a ruler mechanism, thus maintaining the regularity of the spacer size in the CRISPR array. The conservation of the PAM sequence differs between CRISPR-Cas systems and appears to be evolutionarily linked to Cas1 and the leader sequence. New spacers are added to a CRISPR array in a directional manner, occurring preferentially, but not exclusively, adjacent to the leader sequence. Analysis of the type I-E system from ''E. coli'' demonstrated that the first direct repeat adjacent to the leader sequence is copied, with the newly acquired spacer inserted between the first and second direct repeats. The PAM sequence appears to be important during spacer insertion in type I-E systems. That sequence contains a strongly conserved final nucleotide (nt) adjacent to the first nt of the protospacer. This nt becomes the final base in the first direct repeat. This suggests that the spacer acquisition machinery generates single-stranded overhangs in the second-to-last position of the direct repeat and in the PAM during spacer insertion. However, not all CRISPR-Cas systems appear to share this mechanism as PAMs in other organisms do not show the same level of conservation in the final position. It is likely that in those systems, a blunt end is generated at the very end of the direct repeat and the protospacer during acquisition.Insertion variants
Analysis of '' Sulfolobus solfataricus'' CRISPRs revealed further complexities to the canonical model of spacer insertion, as one of its six CRISPR loci inserted new spacers randomly throughout its CRISPR array, as opposed to inserting closest to the leader sequence. Multiple CRISPRs contain many spacers to the same phage. The mechanism that causes this phenomenon was discovered in the type I-E system of ''E. coli''. A significant enhancement in spacer acquisition was detected where spacers already target the phage, even mismatches to the protospacer. This ‘priming’ requires the Cas proteins involved in both acquisition and interference to interact with each other. Newly acquired spacers that result from the priming mechanism are always found on the same strand as the priming spacer. This observation led to the hypothesis that the acquisition machinery slides along the foreign DNA after priming to find a new protospacer.Biogenesis
CRISPR-RNA (crRNA), which later guides the Cas nuclease to the target during the interference step, must be generated from the CRISPR sequence. The crRNA is initially transcribed as part of a single long transcript encompassing much of the CRISPR array. This transcript is then cleaved by Cas proteins to form crRNAs. The mechanism to produce crRNAs differs among CRISPR/Cas systems. In type I-E and type I-F systems, the proteins Cas6e and Cas6f respectively, recognise stem-loops created by the pairing of identical repeats that flank the crRNA. These Cas proteins cleave the longer transcript at the edge of the paired region, leaving a single crRNA along with a small remnant of the paired repeat region. Type III systems also use Cas6, however, their repeats do not produce stem-loops. Cleavage instead occurs by the longer transcript wrapping around the Cas6 to allow cleavage just upstream of the repeat sequence. Type II systems lack the Cas6 gene and instead utilize RNaseIII for cleavage. Functional type II systems encode an extra small RNA that is complementary to the repeat sequence, known as a trans-activating crRNA (tracrRNA). Transcription of the tracrRNA and the primary CRISPR transcript results in base pairing and the formation of dsRNA at the repeat sequence, which is subsequently targeted by RNaseIII to produce crRNAs. Unlike the other two systems, the crRNA does not contain the full spacer, which is instead truncated at one end. CrRNAs associate with Cas proteins to form ribonucleotide complexes that recognize foreign nucleic acids. CrRNAs show no preference between the coding and non-coding strands, which is indicative of an RNA-guided DNA-targeting system. The type I-E complex (commonly referred to as Cascade) requires five Cas proteins bound to a single crRNA.Interference
During the interference stage in type I systems, the PAM sequence is recognized on the crRNA-complementary strand and is required along with crRNA annealing. In type I systems correct base pairing between the crRNA and the protospacer signals a conformational change in Cascade that recruits Cas3 for DNA degradation. Type II systems rely on a single multifunctional protein, Cas9, for the interference step. Cas9 requires both the crRNA and the tracrRNA to function and cleave DNA using its dual HNH and RuvC/RNaseH-like endonuclease domains. Basepairing between the PAM and the phage genome is required in type II systems. However, the PAM is recognized on the same strand as the crRNA (the opposite strand to type I systems). Type III systems, like type I require six or seven Cas proteins binding to crRNAs. The type III systems analysed from ''S. solfataricus'' and ''P. furiosus'' both target the mRNA of phages rather than phage DNA genome, which may make these systems uniquely capable of targeting RNA-based phage genomes. Type III systems were also found to target DNA in addition to RNA using a different Cas protein in the complex, Cas10. The DNA cleavage was shown to be transcription dependent. The mechanism for distinguishing self from foreign DNA during interference is built into the crRNAs and is therefore likely common to all three systems. Throughout the distinctive maturation process of each major type, all crRNAs contain a spacer sequence and some portion of the repeat at one or both ends. It is the partial repeat sequence that prevents the CRISPR-Cas system from targeting the chromosome as base pairing beyond the spacer sequence signals self and prevents DNA cleavage. RNA-guided CRISPR enzymes are classified as type V restriction enzymes.Evolution
The cas genes in the adaptor and effector modules of the CRISPR-Cas system are believed to have evolved from two different ancestral modules. A transposon-like element called casposon encoding the Cas1-like integrase and potentially other components of the adaptation module was inserted next to the ancestral effector module, which likely functioned as an independent innate immune system. The highly conserved cas1 and cas2 genes of the adaptor module evolved from the ancestral module while a variety of class 1 effector cas genes evolved from the ancestral effector module. The evolution of these various class 1 effector module cas genes was guided by various mechanisms, such as duplication events. On the other hand, each type of class 2 effector module arose from subsequent independent insertions of mobile genetic elements. These mobile genetic elements took the place of the multiple gene effector modules to create single gene effector modules that produce large proteins which perform all the necessary tasks of the effector module. The spacer regions of CRISPR-Cas systems are taken directly from foreign mobile genetic elements and thus their long term evolution is hard to trace. The non-random evolution of these spacer regions has been found to be highly dependent on the environment and the particular foreign mobile genetic elements it contains. CRISPR/Cas can immunize bacteria against certain phages and thus halt transmission. For this reason, Koonin described CRISPR/Cas as a Lamarckian inheritance mechanism. However, this was disputed by a critic who noted, "We should remember amarckfor the good he contributed to science, not for things that resemble his theory only superficially. Indeed, thinking of CRISPR and other phenomena as Lamarckian only obscures the simple and elegant way evolution really works". But as more recent studies have been conducted, it has become apparent that the acquired spacer regions of CRISPR-Cas systems are indeed a form of Lamarckian evolution because they are genetic mutations that are acquired and then passed on. On the other hand, the evolution of the Cas gene machinery that facilitates the system evolves through classic Darwinian evolution.Coevolution
Analysis of CRISPR sequences revealed coevolution of host and viral genomes. Cas9 proteins are highly enriched inpathogen
In biology, a pathogen ( el, πάθος, "suffering", "passion" and , "producer of") in the oldest and broadest sense, is any organism or agent that can produce disease. A pathogen may also be referred to as an infectious agent, or simply a ger ...
ic and commensal bacteria. CRISPR/Cas-mediated gene regulation may contribute to the regulation of endogenous bacterial genes, particularly during interaction with eukaryotic hosts. For example, '' Francisella novicida'' uses a unique, small, CRISPR/Cas-associated RNA (scaRNA) to repress an endogenous transcript encoding a bacterial lipoprotein that is critical for ''F. novicida'' to dampen host response and promote virulence.
The basic model of CRISPR evolution is newly incorporated spacers driving phages to mutate their genomes to avoid the bacterial immune response, creating diversity in both the phage and host populations. To resist a phage infection, the sequence of the CRISPR spacer must correspond perfectly to the sequence of the target phage gene. Phages can continue to infect their hosts' given point mutations in the spacer. Similar stringency is required in PAM or the bacterial strain remains phage sensitive.
Rates
A study of 124 ''S. thermophilus'' strains showed that 26% of all spacers were unique and that different CRISPR loci showed different rates of spacer acquisition. Some CRISPR loci evolve more rapidly than others, which allowed the strains' phylogenetic relationships to be determined. A comparative genomic analysis showed that ''E. coli'' and '' S. enterica'' evolve much more slowly than ''S. thermophilus''. The latter's strains that diverged 250 thousand years ago still contained the same spacer complement. Metagenomic analysis of two acid-mine-drainage biofilms showed that one of the analyzed CRISPRs contained extensive deletions and spacer additions versus the other biofilm, suggesting a higher phage activity/prevalence in one community than the other. In the oral cavity, a temporal study determined that 7–22% of spacers were shared over 17 months within an individual while less than 2% were shared across individuals. From the same environment, a single strain was tracked usingPCR PCR or pcr may refer to:
Science
* Phosphocreatine, a phosphorylated creatine molecule
* Principal component regression, a statistical technique
Medicine
* Polymerase chain reaction
** COVID-19 testing, often performed using the polymerase chain r ...
primers specific to its CRISPR system. Broad-level results of spacer presence/absence showed significant diversity. However, this CRISPR added 3 spacers over 17 months, suggesting that even in an environment with significant CRISPR diversity some loci evolve slowly.
CRISPRs were analysed from the metagenomes produced for the human microbiome project. Although most were body-site specific, some within a body site are widely shared among individuals. One of these loci originated from streptococcal species and contained ≈15,000 spacers, 50% of which were unique. Similar to the targeted studies of the oral cavity, some showed little evolution over time.
CRISPR evolution was studied in chemostats using ''S. thermophilus'' to directly examine spacer acquisition rates. In one week, ''S. thermophilus'' strains acquired up to three spacers when challenged with a single phage. During the same interval, the phage developed single nucleotide polymorphisms that became fixed in the population, suggesting that targeting had prevented phage replication absent these mutations.
Another ''S. thermophilus'' experiment showed that phages can infect and replicate in hosts that have only one targeting spacer. Yet another showed that sensitive hosts can exist in environments with high phage titres. The chemostat and observational studies suggest many nuances to CRISPR and phage (co)evolution.
Identification
CRISPRs are widely distributed among bacteria and archaea and show some sequence similarities. Their most notable characteristic is their repeating spacers and direct repeats. This characteristic makes CRISPRs easily identifiable in long sequences of DNA, since the number of repeats decreases the likelihood of a false positive match. Analysis of CRISPRs in metagenomic data is more challenging, as CRISPR loci do not typically assemble, due to their repetitive nature or through strain variation, which confuses assembly algorithms. Where many reference genomes are available,polymerase chain reaction
The polymerase chain reaction (PCR) is a method widely used to rapidly make millions to billions of copies (complete or partial) of a specific DNA sample, allowing scientists to take a very small sample of DNA and amplify it (or a part of it) ...
(PCR) can be used to amplify CRISPR arrays and analyse spacer content. However, this approach yields information only for specifically targeted CRISPRs and for organisms with sufficient representation in public databases to design reliable polymerase chain reaction
The polymerase chain reaction (PCR) is a method widely used to rapidly make millions to billions of copies (complete or partial) of a specific DNA sample, allowing scientists to take a very small sample of DNA and amplify it (or a part of it) ...
(PCR) primers. Degenerate repeat-specific primers can be used to amplify CRISPR spacers directly from environmental samples; amplicons containing two or three spacers can be then computationally assembled to reconstruct long CRISPR arrays.
The alternative is to extract and reconstruct CRISPR arrays from shotgun metagenomic data. This is computationally more difficult, particularly with second generation sequencing technologies (e.g. 454, Illumina), as the short read lengths prevent more than two or three repeat units appearing in a single read. CRISPR identification in raw reads has been achieved using purely ''de novo'' identification or by using direct repeat sequences in partially assembled CRISPR arrays from contigs (overlapping DNA segments that together represent a consensus region of DNA) and direct repeat sequences from published genomes as a hook for identifying direct repeats in individual reads.
Use by phages
Another way for bacteria to defend against phage infection is by having chromosomal islands. A subtype of chromosomal islands called phage-inducible chromosomal island (PICI) is excised from a bacterial chromosome upon phage infection and can inhibit phage replication. PICIs are induced, excised, replicated, and finally packaged into small capsids by certain staphylococcal temperate phages. PICIs use several mechanisms to block phage reproduction. In the first mechanism, PICI-encoded Ppi differentially blocks phage maturation by binding or interacting specifically with phage TerS, hence blocking phage TerS/TerL complex formation responsible for phage DNA packaging. In the second mechanism PICI CpmAB redirects the phage capsid morphogenetic protein to make 95% of SaPI-sized capsid and phage DNA can package only 1/3rd of their genome in these small capsids and hence become nonviable phage. The third mechanism involves two proteins, PtiA and PtiB, that target the LtrC, which is responsible for the production of virion and lysis proteins. This interference mechanism is modulated by a modulatory protein, PtiM, binds to one of the interference-mediating proteins, PtiA, and hence achieves the required level of interference. One study showed that lytic ICP1 phage, which specifically targets '' Vibrio cholerae''serogroup
A serotype or serovar is a distinct variation within a species of bacteria or virus or among immune cells of different individuals. These microorganisms, viruses, or cells are classified together based on their surface antigens, allowing the epi ...
O1, has acquired a CRISPR/Cas system that targets a ''V. cholera'' PICI-like element. The system has 2 CRISPR loci and 9 Cas genes. It seems to be homologous
Homology may refer to:
Sciences
Biology
*Homology (biology), any characteristic of biological organisms that is derived from a common ancestor
*Sequence homology, biological homology between DNA, RNA, or protein sequences
* Homologous chrom ...
to the I-F system found in ''Yersinia pestis
''Yersinia pestis'' (''Y. pestis''; formerly '' Pasteurella pestis'') is a gram-negative, non-motile, coccobacillus bacterium without spores that is related to both '' Yersinia pseudotuberculosis'' and '' Yersinia enterocolitica''. It is a facu ...
''. Moreover, like the bacterial CRISPR/Cas system, ICP1 CRISPR/Cas can acquire new sequences, which allows phage and host to co-evolve.
Certain archaeal viruses were shown to carry mini-CRISPR arrays containing one or two spacers. It has been shown that spacers within the virus-borne CRISPR arrays target other viruses and plasmids, suggesting that mini-CRISPR arrays represent a mechanism of heterotypic superinfection exclusion and participate in interviral conflicts.
Applications
CRISPR gene editing
CRISPR technology has been applied in the food and farming industries to engineer probiotic cultures and to immunize industrial cultures (for yogurt, for instance) against infections. It is also being used in crops to enhance yield, drought tolerance and nutritional value. By the end of 2014, some 1000 research papers had been published that mentioned CRISPR. The technology had been used to functionally inactivate genes in human cell lines and cells, to study '' Candida albicans'', to modify yeasts used to makebiofuels
Biofuel is a fuel that is produced over a short time span from biomass, rather than by the very slow natural processes involved in the formation of fossil fuels, such as oil. According to the United States Energy Information Administration (E ...
, and genetically modify crop strains. Hsu and his colleagues state that the ability to manipulate the genetic sequences allows for reverse engineering that can positively affect biofuel production CRISPR can also be used to change mosquitos so they cannot transmit diseases such as malaria. CRISPR-based approaches utilizing Cas12a have recently been utilized in the successful modification of a broad number of plant species.
In July 2019, CRISPR was used to experimentally treat a patient with a genetic disorder. The patient was a 34-year-old woman with sickle cell disease.
In February 2020, progress was made on HIV
The human immunodeficiency viruses (HIV) are two species of '' Lentivirus'' (a subgroup of retrovirus) that infect humans. Over time, they cause acquired immunodeficiency syndrome (AIDS), a condition in which progressive failure of the immu ...
treatments with 60-80% of the integrated viral DNA removed in mice and some being completely free from the virus after edits involving both LASER ART, a new anti-retroviral therapy, and CRISPR.
In March 2020, CRISPR-modified virus was injected into a patient's eye in an attempt to treat Leber congenital amaurosis.
In the future, CRISPR gene editing could potentially be used to create new species or revive extinct species from closely related ones.
CRISPR-based re-evaluations of claims for gene-disease relationships have led to the discovery of potentially important anomalies.
In July 2021, CRISPR gene editing of hiPSC's was used to study the role of MBNL proteins associated with DM1.
CRISPR as diagnostic tool
 CRISPR associated nucleases have shown to be useful as a tool for molecular testing due to their ability to specifically target nucleic acid sequences in a high background of non-target sequences. In 2016, the Cas9 nuclease was used to deplete unwanted nucleotide sequences in next-generation sequencing libraries while requiring only 250 picograms of initial RNA input. Beginning in 2017, CRISPR associated nucleases were also used for direct diagnostic testing of nucleic acids, down to single molecule sensitivity. CRISPR diversity is used as an analysis target to discern
CRISPR associated nucleases have shown to be useful as a tool for molecular testing due to their ability to specifically target nucleic acid sequences in a high background of non-target sequences. In 2016, the Cas9 nuclease was used to deplete unwanted nucleotide sequences in next-generation sequencing libraries while requiring only 250 picograms of initial RNA input. Beginning in 2017, CRISPR associated nucleases were also used for direct diagnostic testing of nucleic acids, down to single molecule sensitivity. CRISPR diversity is used as an analysis target to discern phylogeny
A phylogenetic tree (also phylogeny or evolutionary tree Felsenstein J. (2004). ''Inferring Phylogenies'' Sinauer Associates: Sunderland, MA.) is a branching diagram or a tree showing the evolutionary relationships among various biological spe ...
and diversity in bacteria, such as in xanthomonad
The Xanthomonadales are a bacterial order within the Gammaproteobacteria. They are one of the largest groups of bacterial phytopathogens, harbouring species such as ''Xanthomonas citri'', ''Xanthomonas euvesicatoria'', ''Xanthomonas oryzae'' and ...
s by Martins ''et al.'', 2019. Early detections of plant pathogens by molecular typing of the pathogen's CRISPRs can be used in agriculture as demonstrated by Shen ''et al.'', 2020.
By coupling CRISPR-based diagnostics to additional enzymatic processes, the detection of molecules beyond nucleic acids is possible. One example of a coupled technology is SHERLOCK-based Profiling of IN vitro Transcription (SPRINT). SPRINT can be used to detect a variety of substances, such as metabolites in patient samples or contaminants in environmental samples, with high throughput or with portable point-of-care devices. CRISPR/Cas platforms are also being explored for detection and inactivation of SARS-CoV-2
Severe acute respiratory syndrome coronavirus 2 (SARS‑CoV‑2) is a strain of coronavirus that causes COVID-19 (coronavirus disease 2019), the respiratory illness responsible for the ongoing COVID-19 pandemic. The virus previously had a No ...
, the virus that causes COVID-19
Coronavirus disease 2019 (COVID-19) is a contagious disease caused by a virus, the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). The first known case was identified in Wuhan, China, in December 2019. The disease quickl ...
. Two different comprehensive diagnostic tests, AIOD-CRISPR and SHERLOCK test have been identified for SARS-CoV-2. The SHERLOCK test is based on a fluorescently labelled press reporter RNA which has the ability to identify 10 copies per microliter. The AIOD-CRISPR helps with robust and highly sensitive visual detection of the viral nucleic acid.
See also
*Anti-CRISPR
Anti-CRISPR (Anti-Clustered Regularly Interspaced Short Palindromic Repeats or Acr) is a group of proteins found in Bacteriophage, phages, that inhibit the normal activity of CRISPR-Cas, the immune system of certain bacteria. CRISPR consists of G ...
* CRISPR/Cas Tools
* CRISPR gene editing
* The CRISPR Journal
* DRACO
* Gene knockout
* Genetics
Genetics is the study of genes, genetic variation, and heredity in organisms.Hartl D, Jones E (2005) It is an important branch in biology because heredity is vital to organisms' evolution. Gregor Mendel, a Moravian Augustinian friar worki ...
* Genome-wide CRISPR-Cas9 knockout screens
* Glossary of genetics
* ''Human Nature'' (2019 documentary film)
* Prime editing Prime editing is a 'search-and-replace' genome editing technology in molecular biology by which the genome of living organisms may be modified. The technology directly writes new genetic information into a targeted DNA site. It uses a fusion protein ...
* RNAi
* SiRNA
* Surveyor nuclease assay
Surveyor nuclease assay is an enzyme mismatch cleavage assay used to detect single base mismatches or small insertions or deletions (indels).
Surveyor nuclease is part of a family of mismatch-specific endonucleases that were discovered in celery ...
* Synthetic biology
* Zinc finger
Notes
References
Further reading
* * * * * * * * * * * * * * * * *External links
* *Protein Data Bank
* * * * * {{DEFAULTSORT:Crispr 1987 in biotechnology 2015 in biotechnology Biological engineering Biotechnology Emerging technologies Genetic engineering Genome editing Jennifer Doudna Molecular biology Non-coding RNA Repetitive DNA sequences Immune system Prokaryote genes