|
Virome
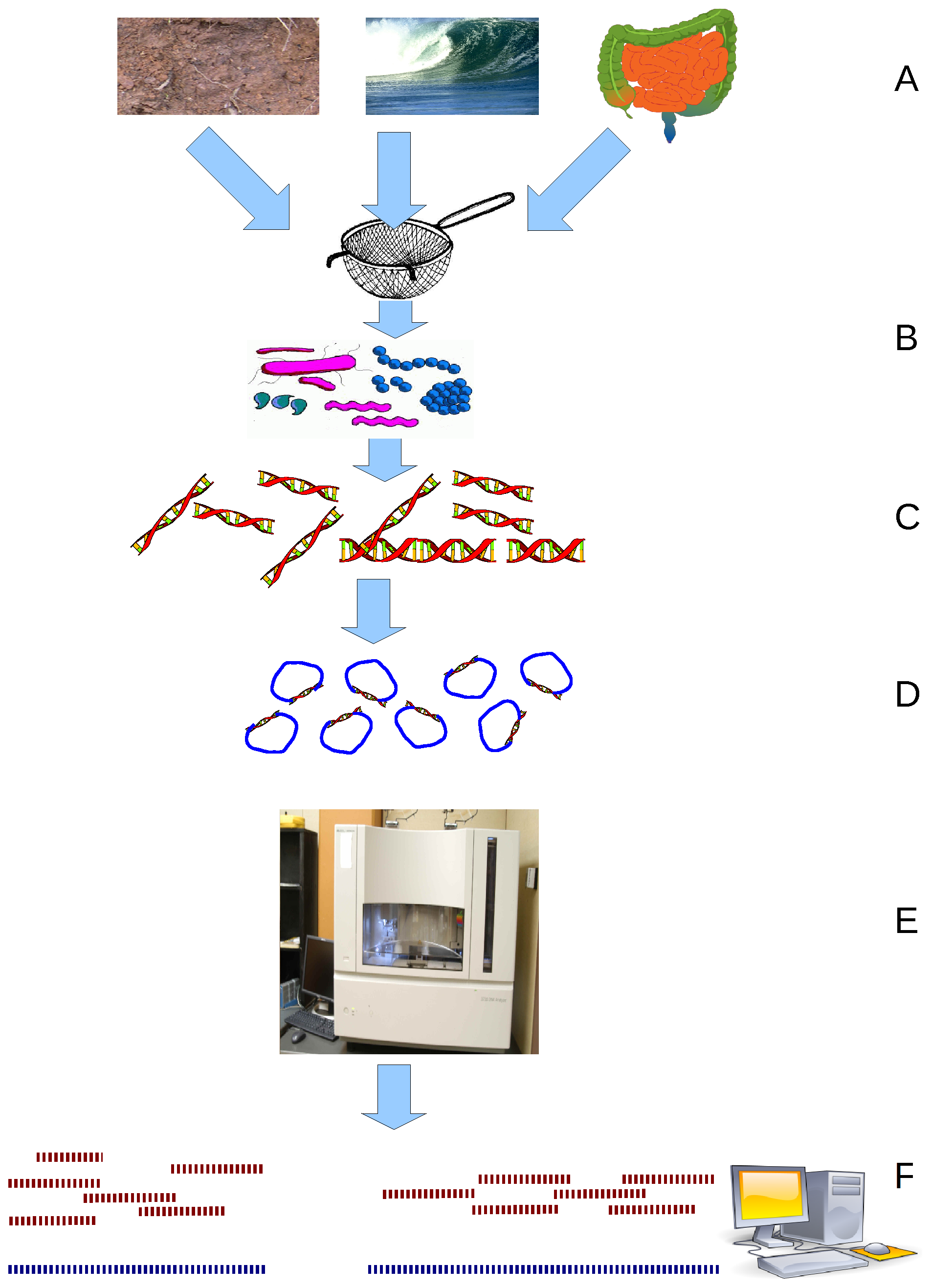
Virome refers to the assemblage of viruses that is often investigated and described by metagenomic sequencing of viral nucleic acids that are found associated with a particular ecosystem, organism or holobiont. The word is frequently used to describe environmental viral shotgun metagenomes. Viruses, including bacteriophages, are found in all environments, and studies of the virome have provided insights into nutrient cycling, development of immunity, and a major source of genes through lysogenic conversion. History The first comprehensive studies of viromes were by shotgun community sequencing, which is frequently referred to as metagenomics. In the 2000s, the Rohwer lab sequenced viromes from seawater, marine sediments, adult human stool, infant human stool, soil, and blood. This group also performed the first RNA virome with collaborators from the Genomic Institute of Singapore. From these early works, it was concluded that most of the genomic diversity is contained in the gl ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Viral Metagenomics
Viral metagenomics is the study of viral genetic material obtained from environmental DNA samples or clinical DNA samples obtained from a Host (biology), host or natural reservoir. Metagenomic methods can be applied to study viruses in any system and has been used to describe various viruses associated with cancerous tumors, extreme environments, terrestrial ecosystems, and the blood and feces of humans. The term virome is also used to refer to viruses investigated by metagenomic sequencing of viral nucleic acids and is frequently used to describe environmental shotgun metagenomes. Viral metagenomics is a culture independent methodology that provides insights on viral diversity, abundance, and functional potential of viruses within the environment. Viruses lack a universal phylogenetic marker making metagenomics the only way to assess the genetic diversity of viruses in an environmental sample . With the advancements of techniques that can exploit next-generation sequencing, virus ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Human Microbiome
The human microbiome is the aggregate of all microbiota that reside on or within human tissues and biofluids along with the corresponding anatomical sites in which they reside, including the skin, mammary glands, seminal fluid, uterus, ovarian follicles, lung, saliva, oral mucosa, conjunctiva, biliary tract, and gastrointestinal tract. Types of human microbiota include bacteria, archaea, fungi, protists and viruses. Though micro-animals can also live on the human body, they are typically excluded from this definition. In the context of genomics, the term ''human microbiome'' is sometimes used to refer to the collective genomes of resident microorganisms; however, the term '' human metagenome'' has the same meaning. Humans are colonized by many microorganisms, with approximately the same order of magnitude of non-human cells as human cells. Some microorganisms that colonize humans are commensal, meaning they co-exist without harming humans; others have a mutualistic relationsh ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Metagenomics
Metagenomics is the study of genetic material recovered directly from environmental or clinical samples by a method called sequencing. The broad field may also be referred to as environmental genomics, ecogenomics, community genomics or microbiomics. While traditional microbiology and microbial genome sequencing and genomics rely upon cultivated clonal cultures, early environmental gene sequencing cloned specific genes (often the 16S rRNA gene) to produce a profile of diversity in a natural sample. Such work revealed that the vast majority of microbial biodiversity had been missed by cultivation-based methods. Because of its ability to reveal the previously hidden diversity of microscopic life, metagenomics offers a powerful lens for viewing the microbial world that has the potential to revolutionize understanding of the entire living world. As the price of DNA sequencing continues to fall, metagenomics now allows microbial ecology to be investigated at a much greater scale ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Mangrove Virome
A mangrove is a shrub or tree that grows in coastal saline or brackish water. The term is also used for tropical coastal vegetation consisting of such species. Mangroves are taxonomically diverse, as a result of convergent evolution in several plant families. They occur worldwide in the tropics and subtropics and even some temperate coastal areas, mainly between latitudes 30° N and 30° S, with the greatest mangrove area within 5° of the equator. Mangrove plant families first appeared during the Late Cretaceous to Paleocene epochs, and became widely distributed in part due to the movement of tectonic plates. The oldest known fossils of mangrove palm date to 75 million years ago. Mangroves are salt-tolerant trees, also called halophytes, and are adapted to live in harsh coastal conditions. They contain a complex salt filtration system and a complex root system to cope with saltwater immersion and wave action. They are adapted to the low-oxygen conditions of waterl ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Metagenomics
Metagenomics is the study of genetic material recovered directly from environmental or clinical samples by a method called sequencing. The broad field may also be referred to as environmental genomics, ecogenomics, community genomics or microbiomics. While traditional microbiology and microbial genome sequencing and genomics rely upon cultivated clonal cultures, early environmental gene sequencing cloned specific genes (often the 16S rRNA gene) to produce a profile of diversity in a natural sample. Such work revealed that the vast majority of microbial biodiversity had been missed by cultivation-based methods. Because of its ability to reveal the previously hidden diversity of microscopic life, metagenomics offers a powerful lens for viewing the microbial world that has the potential to revolutionize understanding of the entire living world. As the price of DNA sequencing continues to fall, metagenomics now allows microbial ecology to be investigated at a much greater scale ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Holobiont
A holobiont is an assemblage of a host and the many other species living in or around it, which together form a discrete ecological unit through symbiosis, though there is controversy over this discreteness. The components of a holobiont are individual species or bionts, while the combined genome of all bionts is the hologenome. The holobiont concept was initially introduced by the German theoretical biologist Adolf Meyer-Abich in 1943, and then apparently independently by Dr. Lynn Margulis in her 1991 book ''Symbiosis as a Source of Evolutionary Innovation''. The concept has evolved since the original formulations. Holobionts include the host, virome, microbiome, and any other organisms which contribute in some way to the functioning of the whole. Well-studied holobionts include reef-building corals and humans. Overview A holobiont is a collection of closely associated species that have complex interactions, such as a plant species and the members of its microbiome. Each sp ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
VIRUS HOST
A virus is a submicroscopic infectious agent that replicates only inside the living cells of an organism. Viruses infect all life forms, from animals and plants to microorganisms, including bacteria and archaea. Since Dmitri Ivanovsky's 1892 article describing a non-bacterial pathogen infecting tobacco plants and the discovery of the tobacco mosaic virus by Martinus Beijerinck in 1898,Dimmock p. 4 more than 9,000 virus species have been described in detail of the millions of types of viruses in the environment. Viruses are found in almost every ecosystem on Earth and are the most numerous type of biological entity. The study of viruses is known as virology, a subspeciality of microbiology. When infected, a host cell is often forced to rapidly produce thousands of copies of the original virus. When not inside an infected cell or in the process of infecting a cell, viruses exist in the form of independent particles, or ''virions'', consisting of (i) the genetic material, i.e. ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Virology
Virology is the Scientific method, scientific study of biological viruses. It is a subfield of microbiology that focuses on their detection, structure, classification and evolution, their methods of infection and exploitation of host (biology), host cell (biology), cells for reproduction, their interaction with host organism physiology and immunity, the diseases they cause, the techniques to isolate and culture them, and their use in research and therapy. The identification of the causative agent of tobacco mosaic disease (TMV) as a novel pathogen by Martinus Beijerinck (1898) is now acknowledged as being the history of virology, official beginning of the field of virology as a discipline distinct from bacteriology. He realized the source was neither a bacterial nor a fungal infection, but something completely different. Beijerinck used the word "virus" to describe the mysterious agent in his 'contagium vivum fluidum' ('contagious living fluid'). Rosalind Franklin proposed the f ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Virus
A virus is a submicroscopic infectious agent that replicates only inside the living cells of an organism. Viruses infect all life forms, from animals and plants to microorganisms, including bacteria and archaea. Since Dmitri Ivanovsky's 1892 article describing a non-bacterial pathogen infecting tobacco plants and the discovery of the tobacco mosaic virus by Martinus Beijerinck in 1898,Dimmock p. 4 more than 9,000 virus species have been described in detail of the millions of types of viruses in the environment. Viruses are found in almost every ecosystem on Earth and are the most numerous type of biological entity. The study of viruses is known as virology, a subspeciality of microbiology. When infected, a host cell is often forced to rapidly produce thousands of copies of the original virus. When not inside an infected cell or in the process of infecting a cell, viruses exist in the form of independent particles, or ''virions'', consisting of (i) the genetic material, i. ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
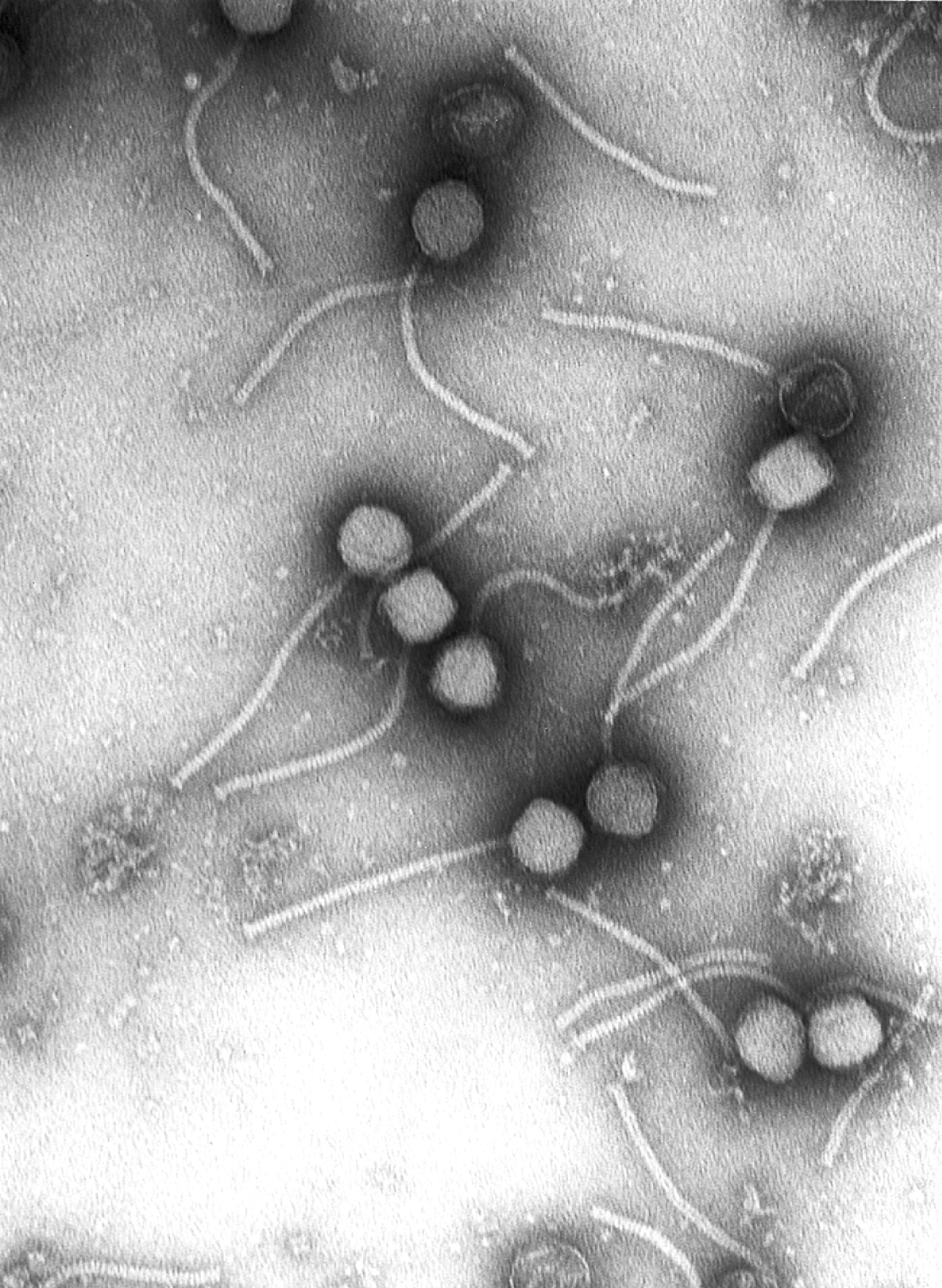
Bacteriophage
A bacteriophage (), also known informally as a ''phage'' (), is a duplodnaviria virus that infects and replicates within bacteria and archaea. The term was derived from "bacteria" and the Greek φαγεῖν ('), meaning "to devour". Bacteriophages are composed of proteins that encapsulate a DNA or RNA genome, and may have structures that are either simple or elaborate. Their genomes may encode as few as four genes (e.g. MS2) and as many as hundreds of genes. Phages replicate within the bacterium following the injection of their genome into its cytoplasm. Bacteriophages are among the most common and diverse entities in the biosphere. Bacteriophages are ubiquitous viruses, found wherever bacteria exist. It is estimated there are more than 1031 bacteriophages on the planet, more than every other organism on Earth, including bacteria, combined. Viruses are the most abundant biological entity in the water column of the world's oceans, and the second largest component of biom ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
CRISPR
CRISPR () (an acronym for clustered regularly interspaced short palindromic repeats) is a family of DNA sequences found in the genomes of prokaryotic organisms such as bacteria and archaea. These sequences are derived from DNA fragments of bacteriophages that had previously infected the prokaryote. They are used to detect and destroy DNA from similar bacteriophages during subsequent infections. Hence these sequences play a key role in the antiviral (i.e. anti-phage) defense system of prokaryotes and provide a form of acquired immunity. CRISPR is found in approximately 50% of sequenced bacterial genomes and nearly 90% of sequenced archaea. Cas9 (or "CRISPR-associated protein 9") is an enzyme that uses CRISPR sequences as a guide to recognize and cleave specific strands of DNA that are complementary to the CRISPR sequence. Cas9 enzymes together with CRISPR sequences form the basis of a technology known as CRISPR-Cas9 that can be used to edit genes within organisms. This editing ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Virosphere
Virosphere (virus diversity, virus world, global virosphere) is the viral part of the biosphere, namely the pool of viruses in all hosts and all environments on planet earth. Virosphere may also refer to a pool of certain group of viruses according to their host - prokaryotic virosphere, archaeal virosphere, Invertebrate virosphere, type of genome - RNA virosphere, dsDNA virosphere or ecological niche - marine virosphere Viral genome diversity The scope of viral genome diversity is enormous compared to cellular life. Cellular life including all known organisms have double stranded DNA genome. Whereas viruses have one of at least 7 different types of genetic information, namely dsDNA, ssDNA, dsRNA, ssRNA+, ssRNA-, ssRNA-RT, dsDNA-RT. Each type of genetic information has its specific manner of mRNA synthesis. Baltimore classification is a system providing overview on these mechanisms for each type of genome. Moreover, in contrast to cellular organisms, viruses don't have uni ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |