Viral metagenomics on:
[Wikipedia]
[Google]
[Amazon]
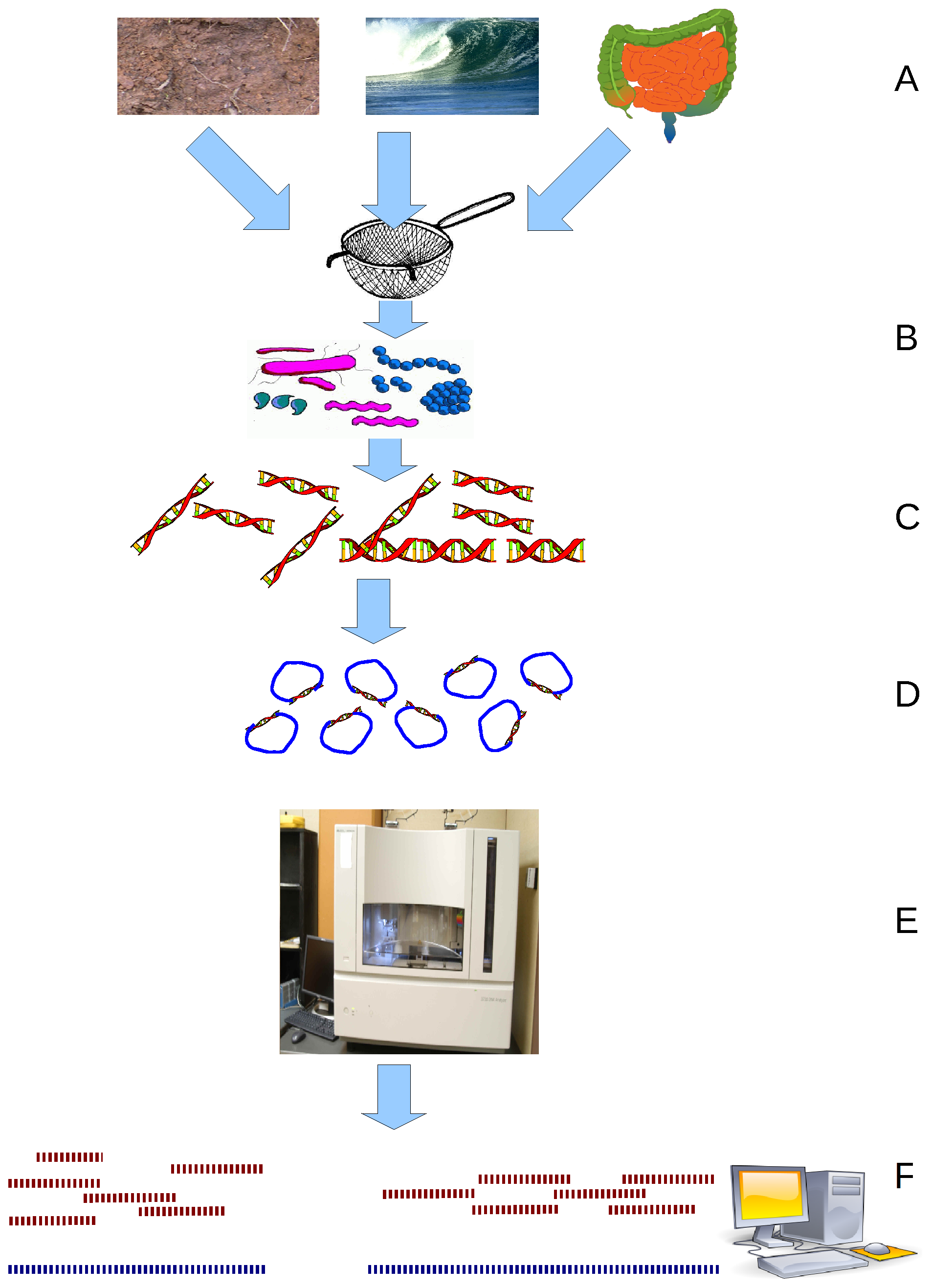 Viral metagenomics is the study of viral genetic material obtained from environmental DNA samples or clinical DNA samples obtained from a
Viral metagenomics is the study of viral genetic material obtained from environmental DNA samples or clinical DNA samples obtained from a
IMG/VR
The IMG Viral Database (IMG/VR).
CAMERA
Cyberinfrastructure for Metagenomics, data repository and tools for metagenomics research.
GOLD
Genomes OnLine Database (GOLD).
IMG/M
The Integrated Microbial Genomes system, for metagenome analysis by the DOE-JGI.
MEGAN
MEtaGenome ANalyzer. A stand-alone metagenome analysis tool.
MetaGeneMark
MetaGeneMark for MetaGenome Gene Finding
Metagenomics and Our Microbial Planet
A website on metagenomics and the vital role of microbes on Earth from th
National Academies.
Metagenomics at the European Bioinformatics Institute
Analysis and archiving of metagenomic data.
Metagenomics: Sequences from the Environment
free ebook from NCBI Bookshelf.
MG-RAST
Metagenomics Rapid Annotation using Subsystem Technology server
The New Science of Metagenomics: Revealing the Secrets of Our Microbial Planet
A report released by the National Research Council in March 2007. Also, see th
Report In Brief.
* http://www.globalviromeproject.org/ Official site of the Global Virome Project * https://www.usaid.gov/ept2 Emerging pandemic threats program EPT-2 * https://www.ecohealthalliance.org/program/predict {{Virus topics Bioinformatics Environmental microbiology Pathogen genomics Metagenomics Microbiology techniques Virology
 Viral metagenomics is the study of viral genetic material obtained from environmental DNA samples or clinical DNA samples obtained from a
Viral metagenomics is the study of viral genetic material obtained from environmental DNA samples or clinical DNA samples obtained from a host
A host is a person responsible for guests at an event or for providing hospitality during it.
Host may also refer to:
Places
* Host, Pennsylvania, a village in Berks County
People
*Jim Host (born 1937), American businessman
* Michel Host ...
or natural reservoir. Metagenomic methods can be applied to study viruses in any system and has been used to describe various viruses associated with cancerous tumors, extreme environments, terrestrial ecosystems, and the blood and feces of humans. The term virome is also used to refer to viruses investigated by metagenomic sequencing of viral nucleic acids and is frequently used to describe environmental shotgun metagenomes. Viral metagenomics is a culture independent methodology that provides insights on viral diversity, abundance, and functional potential of viruses within the environment. Viruses lack a universal phylogenetic marker making metagenomics the only way to assess the genetic diversity of viruses in an environmental sample . With the advancements of techniques that can exploit next-generation sequencing, viruses can now be studied outside of culturable virus-host pairs. This approach has created improvements in molecular epidemiology and accelerated the discovery of novel virus
A virus is a submicroscopic infectious agent that replicates only inside the living cells of an organism. Viruses infect all life forms, from animals and plants to microorganisms, including bacteria and archaea.
Since Dmitri Ivanovsky's ...
es.
History
The term metagenomics was coined in 1998, with the first viral metagenomic study reported a few years later describing uncultured near shore viral communities using shotgun sequencing. The earliest metagenomic studies of viruses were performed using ocean samples, and found that most of the sequenced DNA and RNA viruses had no matches in Virus databases. The researchers also found that previously overlooked ssDNA viruses and prophages are major constituents in some marine environments. Subsequent studies of the soil virome discovered that bacteriophages were equally as prevalent as bacteria in the soil. Acknowledging the importance of viral metagenomics, the International Committee on Taxonomy of Viruses (ICTV) recognizes that genomes assembled from metagenomic data represent a virus and can be classified using the same procedures for viruses isolated via classical virology approaches. The IMG/VR system and the IMG/VR v.2.0 are the largest interactive public virus databases with over 760,000 metagenomic viral sequences and isolate viruses and serves as a starting point for the sequence analysis of viral fragments derived from metagenomic samples.The Global Virome Project
The Global Virome Project (GVP) is an American-led international collaborative research initiative based at the One Health Institute at the University of California, Davis. The project was co-launched by EcoHealth Alliance presidentPeter Daszak
Peter Daszak is a British zoologist, consultant and public expert on disease ecology, in particular on zoonosis. He is the president of EcoHealth Alliance, a nonprofit non-governmental organization that supports various programs on global healt ...
, Nathan Wolfe and Edward Rubin
Edward Rubin is the chief scientific officer at Metabiota, a company that works on epidemic risk and infectious diseases. From 2002 to 2016, he was a researcher at the Lawrence Berkeley National Laboratory and the director of the Department of E ...
of Metabiota
Metabiota is a San Francisco startup that compiles data from around the world to predict disease outbreaks. The company is a partner with USAID's PREDICT (USAID), PREDICT and PREVENT programs. In the early months of the SARS-CoV-2 outbreak, Metab ...
, and former Chinese Center for Disease Control and Prevention
The Chinese Center for Disease Control and Prevention (CCDC; ) is an institution directly under the National Health Commission, based in Changping District, Beijing, China.
Established in 1983, it works to protect public health and safety ...
director George F. Gao.
The goal of the Global Virome Project (GVP) is to identify and prevent future virus outbreaks. The GVP is centered on the massive collection and sequencing of the planet’s unknown viruses, with an estimated 1.6 million viral species yet to be discovered in mammal and bird populations. Of these, 631,000 to 827,000 have zoonotic potential. The cost of identifying these unidentified viruses is a major limitation of the GVP, with a total cost estimate of $1.2 billion. That being said, preventing an outbreak is still less costly than reacting to one, with the total estimated cost of the ongoing COVID-19 pandemic estimated at more than $16 trillion. The Global Virome project also aims to boost infectious disease surveillance around the globe by using low cost sequencing methods in high risk countries to prevent disease outbreaks by expanding on the efforts of the USAID (agency for international development) EPT (Emerging Pandemic Threats) PREDICT
A prediction (Latin ''præ-'', "before," and ''dicere'', "to say"), or forecast, is a statement about a future event or data. They are often, but not always, based upon experience or knowledge. There is no universal agreement about the exact ...
project. The PREDICT project was founded to discover unidentified viral species by sampling animals and humans in countries with high zoonotic disease threat and determine the mechanisms that cause viral spillover into human populations. The Predict project found over 1000 unique viruses in animals and humans.
The Global Virome Project could aid in pandemic surveillance, diagnosis techniques and prevention strategies, and determine the need for pre-emptive production of vaccine and other countermeasures for candidate high-risk viruses. The GVP can also provide further insights into viral pathogenicity and possible biosecurity methods in agriculture.
The Global Virome Project was supposed to be begin sampling wild animal populations in 2020 but were delayed due to the COVID-19 pandemic.
Methods
Direct Metagenomics
Metagenomic analysis uses whole genome shotgun sequencing to characterize microbial diversity in clinical and environmental samples. Total DNA and/or RNA are extracted from the samples and are prepared on a DNA or RNA library for sequencing. These methods have been used to sequence the whole genome of Epstein-Barr virus (EBV) and HCV, however, contaminating nucleic acids can affect the sensitivity to the target viral genome with the proportion of reads related to the target sequence often being low. Due to the uncontrollable nature of environmental DNA samples, the most abundant organisms in the environmental sample are the highest represented in the sequencing data and require large samples to achieve full coverage. That being said, shotgun sequencing ensures that these organisms that would previously go unnoticed in culture dependent methods are represented by some sequence segments. Metagenomics can be used for pathogen discovery or diagnosis with the proper bioinformatic tools and databases that can evaluate the possible pathogen. Metagenomics requires no prior knowledge of the viral genome as it does not require primer or probe design, allowing for rapid response to emerging threats. Because this method is agnostic to expected viral content of a sample, it can be used to identify new virus species or divergent members of known species. It therefore has a role in clinical diagnostics, such as identification of pathogens causing encephalitis or virus-associated cancers.PCR Amplicon Enrichment
PCR amplicon enrichment enriches a portion of the viral genome prior to sequencing. This is done via PCR amplification of primers that are complementary to a known, highly conserved nucleotide sequence. PCR amplicon enrichment is then followed by whole genome sequencing methods and has been used to track the Ebola virus, Zika Virus, and COVID-19 epidemics. PCR amplicon sequencing is more successful for whole genome sequencing of samples with low concentrations. However, with larger viral genomes and the heterogeneity of RNA viruses multiple overlapping primers may be required to cover the amplification of all genotypes. PCR amplicon sequencing requires knowledge of the viral genome prior to sequencing, appropriate primers, and is highly dependent on viral titers, however, PCR amplicon sequencing is a cheaper evaluation method than metagenomic sequencing when studying known viruses with relatively small genomes.Target Enrichment
Target enrichment is a culture independent method that sequences viral genomes directly from clinical sample using small RNA or DNA probes complementary to the pathogens reference sequence. The probes, which can be bound to a solid phase and capture and pull down complementary DNA sequences in the sample. The presence of overlapping probes increases the tolerance for primer mismatches but their design requires high cost and time so a rapid response is limited. DNA capture is followed by brief PCR cycling and shotgun sequencing. Success of this method is dependent available reference sequences to create the probes and is not suitable for characterization of novel viruses. This method has been used to characterize large and small viruses such as HCV,HSV-1
Herpes simplex virus 1 and 2 (HSV-1 and HSV-2), also known by their taxonomical names ''Human alphaherpesvirus 1'' and '' Human alphaherpesvirus 2'', are two members of the human ''Herpesviridae'' family, a set of viruses that produce viral inf ...
, and HCMV.
Applications
Agriculture
Plant viruses pose a global threat to crop production but through metagenomic sequencing and viral database creation, modified plant viruses can be used to aid in plant immunity as well as alter physical appearance. Data obtained on plant virus genomes from metagenomic sequencing can be used to create clone viruses to inoculate the plant with to study viral components and biological characterization of viral agents with increased reproducibility. Engineered mutant virus strains have been used to alter the coloration and size of various ornamental plants and promote the health of crops.Ecology
Viral metagenomics contributes to viral classification without the need of culture based methodologies and has provided vast insights on viral diversity in any system. Metagenomics can be used to study viruses effects on a given ecosystem and how they effect the microbiome as well as monitoring viruses in an ecosystem for possible spillover into human populations. Within the ecosystems, viruses can be studied to determine how they compete with each other as well as viral effects on functions of the host. Viral metagenomics has been used to study unculturable viral communities in marine and soil ecosystems.Infectious Disease Research
Viral metagenomics is readily used to discover novel viruses, with a major focus on those zoonotic or pathogenic to humans. Viral databases obtained from metagenomics provides quick response methods to determine viral infections as well as determine drug resistant variants in clinical samples. The contributions of viral metagenomics to viral classification have aided pandemic surveillance efforts as well as made infectious disease surveillance and testing more affordable. Since the majority of human pandemics are zoonotic in origin, metagenomic surveillance can provide faster identification of novel viruses and their reservoirs.Medicine
Viral metagenomics has been used to test for virus related cancers and difficult to diagnose cases in clinical diagnostics. This method is most often used when conventional and advanced molecular testing cannot find a causative agent for disease. Metagenomic sequencing can also be used to detect pathogenic viruses in clinical samples and provide real time data for a pathogens presence in a population.See also
* MetagenomicsReferences
External links
IMG/VR
The IMG Viral Database (IMG/VR).
CAMERA
Cyberinfrastructure for Metagenomics, data repository and tools for metagenomics research.
GOLD
Genomes OnLine Database (GOLD).
IMG/M
The Integrated Microbial Genomes system, for metagenome analysis by the DOE-JGI.
MEGAN
MEtaGenome ANalyzer. A stand-alone metagenome analysis tool.
MetaGeneMark
MetaGeneMark for MetaGenome Gene Finding
Metagenomics and Our Microbial Planet
A website on metagenomics and the vital role of microbes on Earth from th
National Academies.
Metagenomics at the European Bioinformatics Institute
Analysis and archiving of metagenomic data.
Metagenomics: Sequences from the Environment
free ebook from NCBI Bookshelf.
MG-RAST
Metagenomics Rapid Annotation using Subsystem Technology server
The New Science of Metagenomics: Revealing the Secrets of Our Microbial Planet
A report released by the National Research Council in March 2007. Also, see th
Report In Brief.
* http://www.globalviromeproject.org/ Official site of the Global Virome Project * https://www.usaid.gov/ept2 Emerging pandemic threats program EPT-2 * https://www.ecohealthalliance.org/program/predict {{Virus topics Bioinformatics Environmental microbiology Pathogen genomics Metagenomics Microbiology techniques Virology