|
Viral Fusion Proteins
Membrane fusion proteins (not to be confused with chimeric or fusion proteins) are proteins that cause fusion of biological membranes. Membrane fusion is critical for many biological processes, especially in eukaryotic development and viral entry. Fusion proteins can originate from genes encoded by infectious enveloped viruses, ancient retroviruses integrated into the host genome, or solely by the host genome. Post-transcriptional modifications made to the fusion proteins by the host, namely addition and modification of glycans and acetyl groups, can drastically affect fusogenicity (the ability to fuse). Fusion in eukaryotes Eukaryotic genomes contain several gene families, of host and viral origin, which encode products involved in driving membrane fusion. While adult somatic cells do not typically undergo membrane fusion under normal conditions, gametes and embryonic cells follow developmental pathways to non-spontaneously drive membrane fusion, such as in placental fo ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Fusion Protein
Fusion proteins or chimeric (kī-ˈmir-ik) proteins (literally, made of parts from different sources) are proteins created through the joining of two or more genes that originally coded for separate proteins. Translation of this ''fusion gene'' results in a single or multiple polypeptides with functional properties derived from each of the original proteins. ''Recombinant fusion proteins'' are created artificially by recombinant DNA technology for use in biological research or therapeutics. '' Chimeric'' or ''chimera'' usually designate hybrid proteins made of polypeptides having different functions or physico-chemical patterns. ''Chimeric mutant proteins'' occur naturally when a complex mutation, such as a chromosomal translocation, tandem duplication, or retrotransposition creates a novel coding sequence containing parts of the coding sequences from two different genes. Naturally occurring fusion proteins are commonly found in cancer cells, where they may function as oncoproteins ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Nervous System
In biology, the nervous system is the highly complex part of an animal that coordinates its actions and sensory information by transmitting signals to and from different parts of its body. The nervous system detects environmental changes that impact the body, then works in tandem with the endocrine system to respond to such events. Nervous tissue first arose in wormlike organisms about 550 to 600 million years ago. In vertebrates it consists of two main parts, the central nervous system (CNS) and the peripheral nervous system (PNS). The CNS consists of the brain and spinal cord. The PNS consists mainly of nerves, which are enclosed bundles of the long fibers or axons, that connect the CNS to every other part of the body. Nerves that transmit signals from the brain are called motor nerves or '' efferent'' nerves, while those nerves that transmit information from the body to the CNS are called sensory nerves or '' afferent''. Spinal nerves are mixed nerves that serve both fu ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
ERV3
HERV-R_7q21.2 provirus ancestral envelope (Env) polyprotein is a protein that in humans is encoded by the ''ERV3'' gene. Function The human genome includes many retroelements including the human endogenous retroviruses (HERVs), which compose about 7-8% of the human genome. ERV3, one of the most studied HERVs, is thought to have integrated 30 to 40 million years ago and is present in higher primates with the exception of gorillas. Taken together, the observation of genome conservation, the detection of transcript expression, and the presence of conserved ORFs is circumstantial evidence for a functional role. Similar endogenous retroviral Env genes like syncytin-1 have important roles in placental formation and embryonic development by enabling cell-cell fusion. Despite its origin as an Env gene, ERV3 has a premature stop codon that precludes any cell-cell fusion functionality. However, it does have an immunosuppressive function that helps the fetus evade a damaging maternal imm ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Syncytin-2
Syncytin-2 also known as endogenous retrovirus group FRD member 1 is a protein that in humans is encoded by the ERVFRD-1 gene. This protein plays a key role in the implantation of human embryos in the womb. This gene is conserved among all primates, with an estimated age of 45 million years. The receptor for this fusogenic ''env'' protein is MFSD2. The mouse syncytins are not true orthologues. The virus, along with some very similar insertions, belong to a group under the ''Gammaretrovirus''-like class I ERVs. Similar ERVs are found in artiodactyl The even-toed ungulates (Artiodactyla , ) are ungulates—hoofed animals—which bear weight equally on two (an even number) of their five toes: the third and fourth. The other three toes are either present, absent, vestigial, or pointing poster ...s, a result of an independent integration event. A proposed nomenclature suggests putting all such "class I" elements in a genus-level taxon separate from ''Gammaretrovirus''. Reference ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Syncytin-1
Syncytin-1 also known as enverin is a protein found in humans and other primates that is encoded by the ERVW-1 gene ( endogenous retrovirus group W envelope member 1). Syncytin-1 is a cell-cell fusion protein whose function is best characterized in placental development. The placenta in turn aids in embryo attachment to the uterus and establishment of a nutrient supply. The gene encoding this protein is an endogenous retroviral element that is the remnant of an ancient retroviral infection integrated into the primate germ line. In the case of syncytin-1 (which is found in humans, apes, and Old World but not New World monkeys), this integration likely occurred more than 25 million years ago. Syncytin-1 is one of two known syncytin proteins expressed in catarrhini primates (the other being syncytin-2) and one of many viral genomes incorporated on multiple occasions over evolutionary time in diverse mammalian species. ERVW-1 is located within ERVWE1, a full length provirus on ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
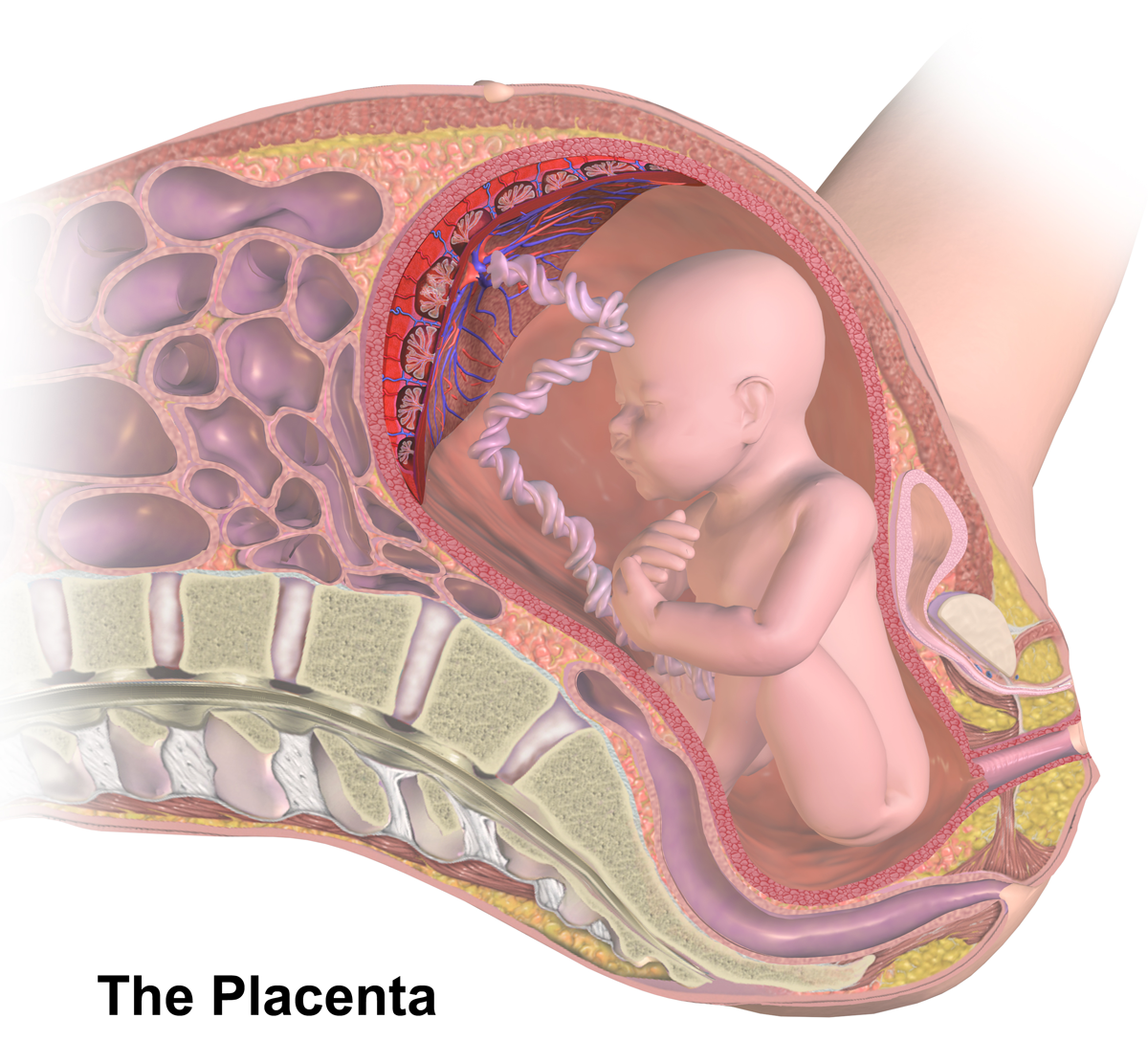
Placenta
The placenta is a temporary embryonic and later fetal organ that begins developing from the blastocyst shortly after implantation. It plays critical roles in facilitating nutrient, gas and waste exchange between the physically separate maternal and fetal circulations, and is an important endocrine organ, producing hormones that regulate both maternal and fetal physiology during pregnancy. The placenta connects to the fetus via the umbilical cord, and on the opposite aspect to the maternal uterus in a species-dependent manner. In humans, a thin layer of maternal decidual (endometrial) tissue comes away with the placenta when it is expelled from the uterus following birth (sometimes incorrectly referred to as the 'maternal part' of the placenta). Placentas are a defining characteristic of placental mammals, but are also found in marsupials and some non-mammals with varying levels of development. Mammalian placentas probably first evolved about 150 million to 200 million years ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Syncytiotrophoblast
Syncytiotrophoblast (from the Greek 'syn'- "together"; 'cytio'- "of cells"; 'tropho'- "nutrition"; 'blast'- "bud") is the epithelial covering of the highly vascular embryonic placental villi, which invades the wall of the uterus to establish nutrient circulation between the embryo and the mother. It is a multi-nucleate, terminally differentiated syncytium, extending to 13cm. Function It is the outer layer of the trophoblasts and actively invades the uterine wall, during implantation, rupturing maternal capillaries and thus establishing an interface between maternal blood and embryonic extracellular fluid, facilitating passive exchange of material between the mother and the embryo. The syncytial property is important since the mother's immune system includes white blood cells that are able to migrate into tissues by "squeezing" in between cells. If they were to reach the fetal side of the placenta many foreign proteins would be recognised, triggering an immune reaction. Howeve ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Endogenous Retroviruses
Endogenous retroviruses (ERVs) are endogenous viral elements in the genome that closely resemble and can be derived from retroviruses. They are abundant in the genomes of jawed vertebrates, and they comprise up to 5–8% of the human genome (lower estimates of ~1%). ERVs are a vertically inherited proviral sequence and a subclass of a type of gene called a transposon, which can normally be packaged and moved within the genome to serve a vital role in gene expression and in regulation. ERVs however lack most transposon functions, are typically not infectious and are often defective genomic remnants of the retroviral replication cycle. They are distinguished as germline provirus retroelements due to their integration and reverse-transcription into the nuclear genome of the host cell. Researchers have suggested that retroviruses evolved from a type of transposon called a retrotransposon, a Class I element; these genes can mutate and instead of moving to another location in the ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Env (gene)
''Env'' is a viral gene that encodes the protein forming the viral envelope. The expression of the ''env'' gene enables retroviruses to target and attach to specific cell types, and to infiltrate the target cell membrane. Analysis of the structure and sequence of several different ''env'' genes suggests that Env proteins are type 1 fusion machines. Type 1 fusion machines initially bind a receptor on the target cell surface, which triggers a conformational change, allowing for binding of the fusion protein. The fusion peptide inserts itself in the host cell membrane and brings the host cell membrane very close to the viral membrane to facilitate membrane fusion. While there are significant differences in sequence of the ''env'' gene between retroviruses, the gene is always located downstream of ''gag'', ''pro'', and '' pol''. The ''env'' mRNA must be spliced for expression. The mature product of the ''env'' gene is the viral spike protein, which has two main parts: the surface pro ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Vesicle-associated Membrane Protein
Vesicle associated membrane proteins (VAMP) are a family of SNARE proteins with similar structure, and are mostly involved in vesicle fusion. * VAMP1 and VAMP2 proteins known as synaptobrevins are expressed in brain and are constituents of the synaptic vesicles, where they participate in neurotransmitter release. * VAMP3 (known as cellubrevin) is ubiquitously expressed and participates in regulated and constitutive exocytosis as a constituent of secretory granules and secretory vesicles. * VAMP5 and VAMP7 ( SYBL1) participate in constitutive exocytosis. ** VAMP5 is a constituent of secretory vesicles, myotubes and tubulovesicular structures. ** VAMP7 is found both in secretory granules and endosomes. * VAMP8 (known as endobrevin) participates in endocytosis and is found in early endosomes. VAMP8 also participates the regulated exocytosis in pancreatic acinar cells. *VAMP4 Vesicle-associated membrane protein 4 is a protein that in humans is encoded by the ''VAMP4'' gene. ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Heimdallarchaeota
Asgard or Asgardarchaeota is a proposed superphylum consisting of a group of archaea that includes Lokiarchaeota, Thorarchaeota, Odinarchaeota, and Heimdallarchaeota. It appears the eukaryotes emerged within the Asgard, in a branch containing the Heimdallarchaeota. This supports the two-domain system of classification over the three-domain system. Discovery and nomenclature In the summer of 2010, sediments were analysed from a gravity core taken in the rift valley on the Knipovich ridge in the Arctic Ocean, near the Loki's Castle hydrothermal vent site. Specific sediment horizons previously shown to contain high abundances of novel archaeal lineages were subjected to metagenomic analysis. In 2015, an Uppsala University-led team proposed the Lokiarchaeota phylum based on phylogenetic analyses using a set of highly conserved protein-coding genes. Through a reference to the hydrothermal vent complex from which the first genome sample originated, the name refers to Loki, the Nors ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Archaeal
Archaea ( ; singular archaeon ) is a domain of single-celled organisms. These microorganisms lack cell nuclei and are therefore prokaryotes. Archaea were initially classified as bacteria, receiving the name archaebacteria (in the Archaebacteria kingdom), but this term has fallen out of use. Archaeal cells have unique properties separating them from the other two domains, Bacteria and Eukaryota. Archaea are further divided into multiple recognized phyla. Classification is difficult because most have not been isolated in a laboratory and have been detected only by their gene sequences in environmental samples. Archaea and bacteria are generally similar in size and shape, although a few archaea have very different shapes, such as the flat, square cells of ''Haloquadratum walsbyi''. Despite this morphological similarity to bacteria, archaea possess genes and several metabolic pathways that are more closely related to those of eukaryotes, notably for the enzymes involved in ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |



.jpg)