|
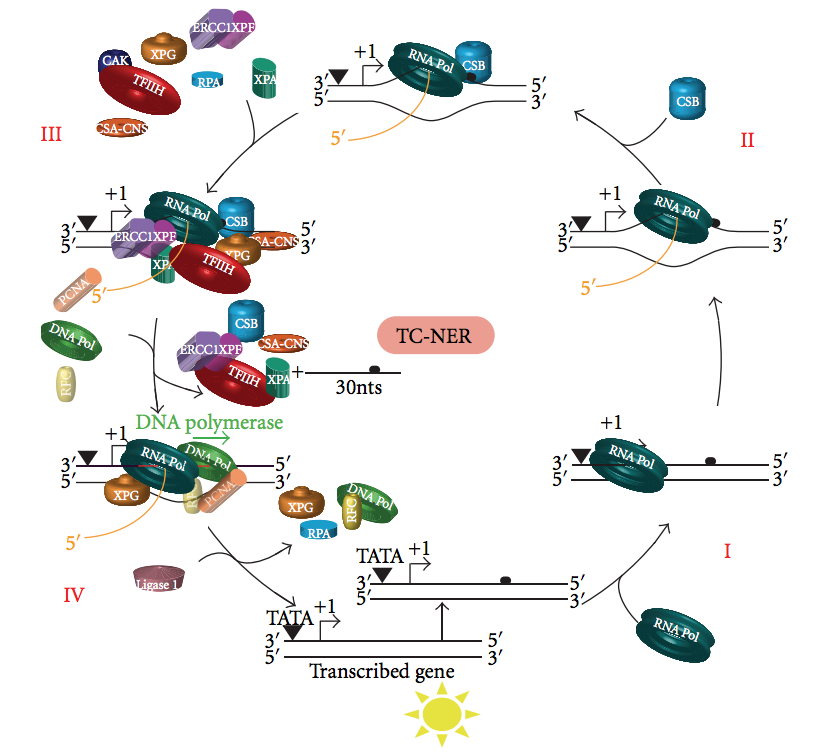
Transcription-coupled Nucleotide Excision Repair
Nucleotide excision repair is a DNA repair mechanism. DNA damage occurs constantly because of chemicals (e.g. intercalating agents), radiation and other mutagens. Three excision repair pathways exist to repair single stranded DNA damage: Nucleotide excision repair (NER), base excision repair (BER), and DNA mismatch repair (MMR). While the BER pathway can recognize specific non-bulky lesions in DNA, it can correct only damaged bases that are removed by specific glycosylases. Similarly, the MMR pathway only targets mismatched Watson-Crick base pairs. Nucleotide excision repair (NER) is a particularly important excision mechanism that removes DNA damage induced by ultraviolet light (UV). UV DNA damage results in bulky DNA adducts - these adducts are mostly thymine dimers and 6,4-photoproducts. Recognition of the damage leads to removal of a short single-stranded DNA segment that contains the lesion. The undamaged single-stranded DNA remains and DNA polymerase uses it as a temp ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Nucleotide Excision Repair-journal
Nucleotides are organic molecules consisting of a nucleoside and a phosphate. They serve as monomeric units of the nucleic acid polymers – deoxyribonucleic acid (DNA) and ribonucleic acid (RNA), both of which are essential biomolecules within all life-forms on Earth. Nucleotides are obtained in the diet and are also synthesized from common nutrients by the liver. Nucleotides are composed of three subunit molecules: a nucleobase, a five-carbon sugar (ribose or deoxyribose), and a phosphate group consisting of one to three phosphates. The four nucleobases in DNA are guanine, adenine, cytosine and thymine; in RNA, uracil is used in place of thymine. Nucleotides also play a central role in metabolism at a fundamental, cellular level. They provide chemical energy—in the form of the nucleoside triphosphates, adenosine triphosphate (ATP), guanosine triphosphate (GTP), cytidine triphosphate (CTP) and uridine triphosphate (UTP)—throughout the cell for the many cellular functio ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Eukaryotes
Eukaryotes () are organisms whose cells have a nucleus. All animals, plants, fungi, and many unicellular organisms, are Eukaryotes. They belong to the group of organisms Eukaryota or Eukarya, which is one of the three domains of life. Bacteria and Archaea (both prokaryotes) make up the other two domains. The eukaryotes are usually now regarded as having emerged in the Archaea or as a sister of the Asgard archaea. This implies that there are only two domains of life, Bacteria and Archaea, with eukaryotes incorporated among archaea. Eukaryotes represent a small minority of the number of organisms, but, due to their generally much larger size, their collective global biomass is estimated to be about equal to that of prokaryotes. Eukaryotes emerged approximately 2.3–1.8 billion years ago, during the Proterozoic eon, likely as flagellated phagotrophs. Their name comes from the Greek εὖ (''eu'', "well" or "good") and κάρυον (''karyon'', "nut" or "kernel"). Eu ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Transcription Coupled Repair
Nucleotide excision repair is a DNA repair mechanism. DNA damage occurs constantly because of chemicals (e.g. intercalating agents), radiation and other mutagens. Three excision repair pathways exist to repair single stranded DNA damage: Nucleotide excision repair (NER), base excision repair (BER), and DNA mismatch repair (MMR). While the BER pathway can recognize specific non-bulky lesions in DNA, it can correct only damaged bases that are removed by specific glycosylases. Similarly, the MMR pathway only targets mismatched Watson-Crick base pairs. Nucleotide excision repair (NER) is a particularly important excision mechanism that removes DNA damage induced by ultraviolet light (UV). UV DNA damage results in bulky DNA adducts - these adducts are mostly thymine dimers and 6,4-photoproducts. Recognition of the damage leads to removal of a short single-stranded DNA segment that contains the lesion. The undamaged single-stranded DNA remains and DNA polymerase uses it as a templat ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Global Genomic Repair
Global means of or referring to a globe and may also refer to: Entertainment * ''Global'' (Paul van Dyk album), 2003 * ''Global'' (Bunji Garlin album), 2007 * ''Global'' (Humanoid album), 1989 * ''Global'' (Todd Rundgren album), 2015 * Bruno J. Global, a character in the anime series ''The Super Dimension Fortress Macross'' Companies and brands Television * Global Television Network, in Canada ** Global BC, on-air brand of CHAN-TV, a television station in Vancouver, British Columbia, Canada ** Global Okanagan, on-air brand of CHBC-TV, a television station in Kelowna, British Columbia, Canada ** Global Toronto, a television station in Toronto ** Global Edmonton ** Global Calgary ** Global Montreal ** Global Maritimes ** Canwest Global, former parent company of Global Television Network * Global TV (Venezuela), a regional channel in Venezuela Other industries * Global (cutlery), a Japanese brand * Global Aviation Holdings, the parent company of World Airways, Inc., and North Am ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Nucleotide Excision Repair
Nucleotide excision repair is a DNA repair mechanism. DNA damage occurs constantly because of chemicals (e.g. intercalating agents), radiation and other mutagens. Three excision repair pathways exist to repair single stranded DNA damage: Nucleotide excision repair (NER), base excision repair (BER), and DNA mismatch repair (MMR). While the BER pathway can recognize specific non-bulky lesions in DNA, it can correct only damaged bases that are removed by specific glycosylases. Similarly, the MMR pathway only targets mismatched Watson-Crick base pairs. Nucleotide excision repair (NER) is a particularly important excision mechanism that removes DNA damage induced by ultraviolet light (UV). UV DNA damage results in bulky DNA adducts - these adducts are mostly thymine dimers and 6,4-photoproducts. Recognition of the damage leads to removal of a short single-stranded DNA segment that contains the lesion. The undamaged single-stranded DNA remains and DNA polymerase uses it as a templa ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
RAD23B
UV excision repair protein RAD23 homolog B is a protein that in humans is encoded by the ''RAD23B'' gene. Function The protein encoded by this gene is one of two human homologs of Saccharomyces cerevisiae Rad23, a protein involved in nucleotide excision repair (NER). This protein was found to be a component of the protein complex that specifically complements the NER defect of xeroderma pigmentosum group C (XP-c) cell extracts in vitro. This protein was also shown to interact with, and elevate the nucleotide excision activity of 3-methyladenine-DNA glycosylase (MPG), which suggested a role in DNA damage recognition in base excision repair. This protein contains an N-terminal ubiquitin-like domain, which was reported to interact with 26S proteasome, and thus this protein may be involved in the ubiquitin mediated proteolytic pathway in cells. Role in DNA repair The complex of XPC-RAD23B is the initial damage recognition factor in global genomic nucleotide excision repair (GG-NER) ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
RAD23A
UV excision repair protein RAD23 homolog A is a protein that in humans is encoded by the ''RAD23A'' gene. Function The protein encoded by this gene is one of two human homologs of ''Saccharomyces cerevisiae'' Rad23, a protein involved in nucleotide excision repair (NER). This protein was shown to interact with, and elevate the nucleotide excision activity of 3-methyladenine-DNA glycosylase (MPG), which suggested a role in DNA damage recognition in base excision repair. This protein contains an N-terminal ubiquitin-like domain, which was reported to interact with 26S proteasome, as well as with ubiquitin protein ligase E6AP, and thus suggests that this protein may be involved in the ubiquitin mediated proteolytic pathway in cells. Interactions RAD23A has been shown to interact with: * Ataxin 3, * PSMD4, and * Sequestosome 1 Sequestosome-1 is a protein that in humans is encoded by the ''SQSTM1'' gene. Also known as the ubiquitin-binding protein p62, it is an autophag ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Replication Protein A
Replication protein A (RPA) is the major protein that binds to single-stranded DNA (ssDNA) in eukaryotic cells. In vitro, RPA shows a much higher affinity for ssDNA than RNA or double-stranded DNA. RPA is required in replication, recombination and repair processes such as nucleotide excision repair and homologous recombination. It also plays roles in responding to damaged DNA. Structure RPA is a heterotrimer, composed of the subunits RPA1 (RPA70) (70kDa subunit), RPA2 (RPA32) (32kDa subunit) and RPA3 (RPA14) (14kDa subunit). The three RPA subunits contain six OB-folds (oligonucleotide/oligosaccharide binding),with DNA-binding domains (DBD) designated DBDs A-F, that bind RPA to single-stranded DNA. DBDs A, B, C and F are located on RPA1, DBD D is located on RPA2, and DBD E is located on RPA3. DBDs C, D, and E make up the trimerization core of the protein with flexible linker regions connecting them all together. Due to these flexible linker regions RPA is considere ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
ERCC1
DNA excision repair protein ERCC-1 is a protein that in humans is encoded by the ''ERCC1'' gene. Together with ERCC4, ERCC1 forms the ERCC1-XPF enzyme complex that participates in DNA repair and DNA recombination. Many aspects of these two gene products are described together here because they are partners during DNA repair. The ERCC1-XPF nuclease is an essential activity in the pathway of DNA nucleotide excision repair (NER). The ERCC1-XPF nuclease also functions in pathways to repair double-strand breaks in DNA, and in the repair of “crosslink” damage that harmfully links the two DNA strands. Cells with disabling mutations in ''ERCC1'' are more sensitive than normal to particular DNA damaging agents, including ultraviolet (UV) radiation and to chemicals that cause crosslinking between DNA strands. Genetically engineered mice with disabling mutations in ERCC1 have defects in DNA repair, accompanied by metabolic stress-induced changes in physiology that result in premature a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
ERCC6
DNA excision repair protein ERCC-6 (also CS-B protein) is a protein that in humans is encoded by the ''ERCC6'' gene. The ''ERCC6'' gene is located on the long arm of chromosome 10 at position 11.23.NIH. "ERCC6 Gene." Genetics Home Reference. National Institutes of Health, 16 Feb. 2015. Web. 22 Feb. 2015. . Having 1 or more copies of a mutated ERCC6 causes Cockayne syndrome, type II. Function DNA can be damaged by ultraviolet radiation, toxins, radioactive substances, and reactive biochemical intermediates like free radicals. The ERCC6 protein is involved in repairing the genome when specific genes undergoing transcription (dubbed ''active genes'') are inoperative; as such, ERCC6 serves as a transcription-coupled excision repair protein, being one of the fundamental enzymes in active gene repair. Structure and Mechanism CSB has been found to exhibit ATPase properties; there are contradictory publications regarding the effect of ATP concentration on CSB's activity. The most ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
ERCC8
DNA excision repair protein ERCC-8 is a protein that in humans is encoded by the ''ERCC8'' gene. This gene encodes a WD repeat protein, which interacts with the Cockayne syndrome type B (CSB) and p44 proteins, the latter being a subunit of the RNA polymerase II transcription factor II H. Mutations in this gene have been identified in patients with the hereditary disease Cockayne syndrome (CS). CS is an accelerated aging disorder characterized by photosensitivity, impaired development and multi-system progressive degeneration. The CS cells are abnormally sensitive to ultraviolet radiation and are defective in the repair of transcriptionally active genes. Multiple alternatively spliced transcript variants encoding different isoforms have been found for this gene. CS arises from germline mutations in either of two genes ''CSA(ERCC8)'' or ''CSB(ERCC6)''. ''CSA'' mutations generally give rise to a more moderate form of CS than ''CSB'' mutations. Mutations in the ''CSA'' gene accou ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
XPG-Nterminus
In molecular biology the protein domain XPG refers to, in this case, the N-terminus of XPG. The XPG protein can be corrected by a 133 kDa nuclear protein, XPGC. XPGC is an acidic protein that confers normal ultraviolet (UV) light resistance. It is a magnesium-dependent, single-strand DNA endonuclease that makes structure-specific endonucleolytic incisions in a DNA substrate containing a duplex region and single-stranded arms. XPGC cleaves one strand of the duplex at the border with the single-stranded region. Homology XPG belongs to a family of proteins that includes: * RAD2 from ''Saccharomyces cerevisiae'' (Baker's yeast) and rad13 from ''Schizosaccharomyces pombe'' (Fission yeast), which are single-stranded DNA endonucleases,; * mouse and human FEN-1, a structure-specific endonuclease; * RAD2 from fission yeast and RAD27 from budding yeast; * fission yeast exo1, a 5' -3' double-stranded DNA exonuclease that may act in a pathway that corrects mismatched base pairs; * y ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |