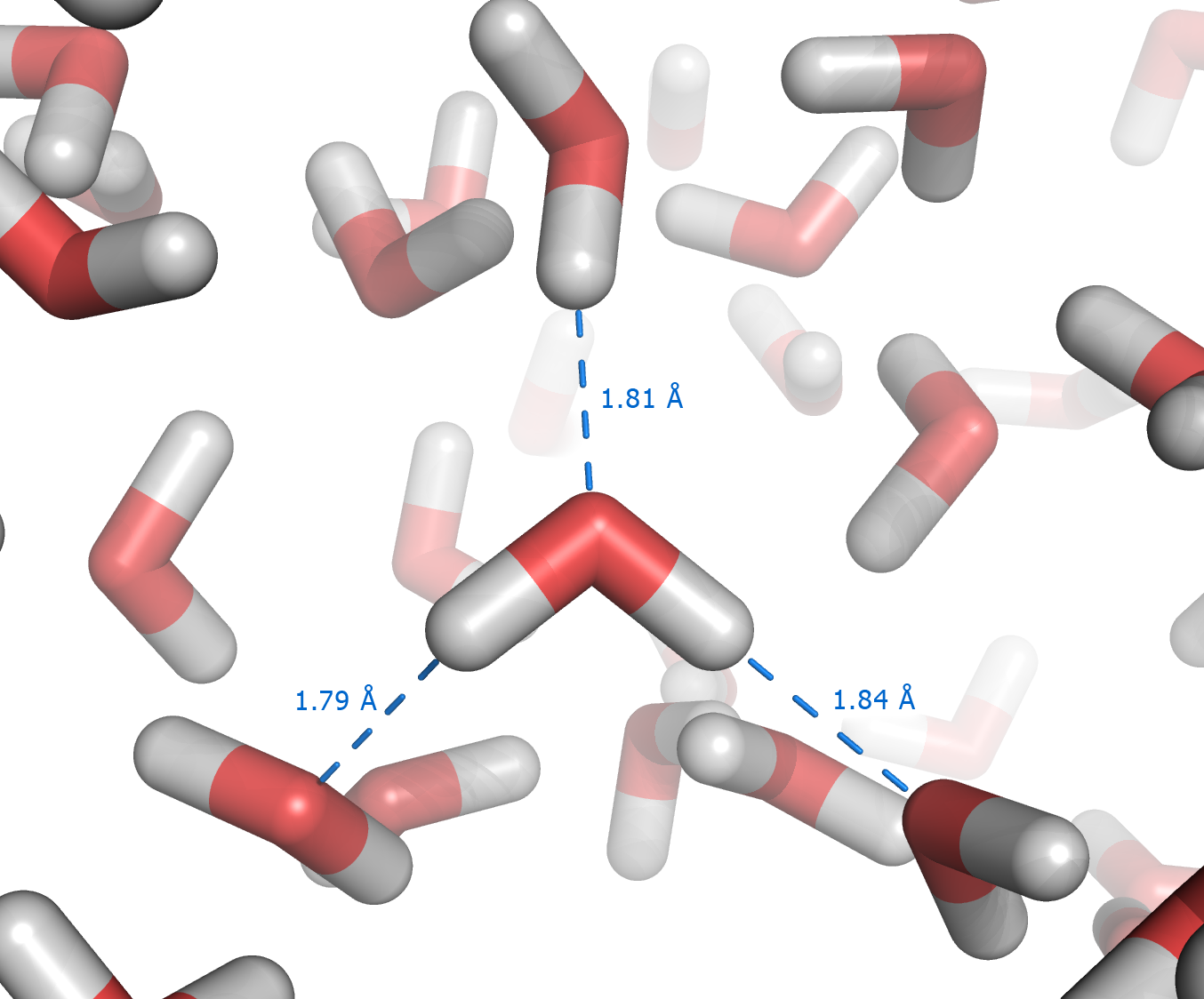
|
Sodium-calcium Exchanger
The sodium-calcium exchanger (often denoted Na+/Ca2+ exchanger, exchange protein, or NCX) is an antiporter membrane protein that removes calcium from cells. It uses the energy that is stored in the electrochemical gradient of sodium (Na+) by allowing Na+ to flow down its gradient across the plasma membrane in exchange for the countertransport of calcium ions (Ca2+). A single calcium ion is exported for the import of three sodium ions. The exchanger exists in many different cell types and animal species. The NCX is considered one of the most important cellular mechanisms for removing Ca2+. The exchanger is usually found in the plasma membranes and the mitochondria and endoplasmic reticulum of excitable cells. Function The sodium–calcium exchanger is only one of the systems by which the cytoplasmic concentration of calcium ions in the cell is kept low. The exchanger does not bind very tightly to Ca2+ (has a low affinity), but it can transport the ions rapidly (has a high cap ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Antiporter
An antiporter (also called exchanger or counter-transporter) is a cotransporter and integral membrane protein involved in secondary active transport of two or more different molecules or ions across a phospholipid membrane such as the plasma membrane in opposite directions, one into the cell and one out of the cell. Na+/H+ antiporters have been reviewed. In secondary active transport, one species of solute moves along its electrochemical gradient, allowing a different species to move against its own electrochemical gradient. This movement is in contrast to primary active transport, in which all solutes are moved against their concentration gradients, fueled by ATP. Transport may involve one or more of each type of solute. For example, the Na+/Ca2+ exchanger, found in the plasma membrane of many cells, moves three sodium ions in one direction, and one calcium ion in the other. Role in Homeostatic Mechanisms Na+/H+ Antiporters Antiporters, such as Na+/H+ antiporter protein ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Excitation-contraction Coupling
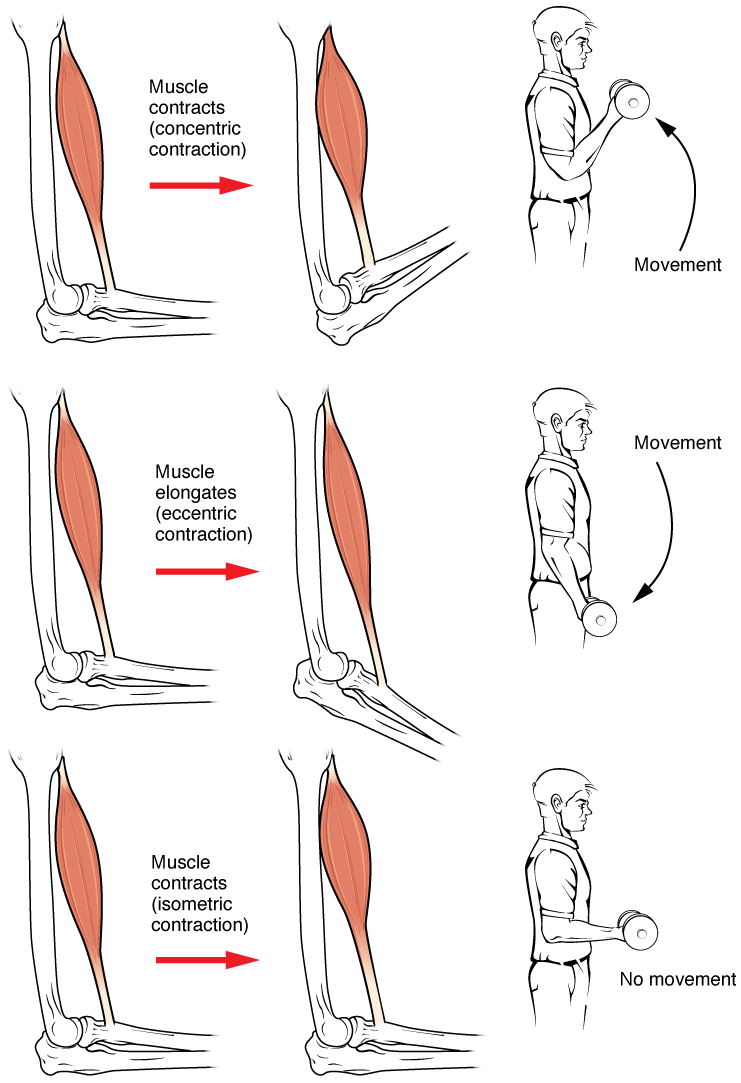
Muscle contraction is the activation of tension-generating sites within muscle cells. In physiology, muscle contraction does not necessarily mean muscle shortening because muscle tension can be produced without changes in muscle length, such as when holding something heavy in the same position. The termination of muscle contraction is followed by muscle relaxation, which is a return of the muscle fibers to their low tension-generating state. For the contractions to happen, the muscle cells must rely on the interaction of two types of filaments which are the thin and thick filaments. Thin filaments are two strands of actin coiled around each, and thick filaments consist of mostly elongated proteins called myosin. Together, these two filaments form myofibrils which are important organelles in the skeletal muscle system. Muscle contraction can also be described based on two variables: length and tension. A muscle contraction is described as isometric if the muscle tension changes ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Dimer
In biochemistry, a protein dimer is a macromolecular complex formed by two protein monomers, or single proteins, which are usually non-covalently bound. Many macromolecules, such as proteins or nucleic acids, form dimers. The word ''dimer'' has roots meaning "two parts", '' di-'' + '' -mer''. A protein dimer is a type of protein quaternary structure. A protein homodimer is formed by two identical proteins. A protein heterodimer is formed by two different proteins. Most protein dimers in biochemistry are not connected by covalent bonds. An example of a non-covalent heterodimer is the enzyme reverse transcriptase, which is composed of two different amino acid chains. An exception is dimers that are linked by disulfide bridges such as the homodimeric protein NEMO. Some proteins contain specialized domains to ensure dimerization (dimerization domains) and specificity. The G protein-coupled cannabinoid receptors have the ability to form both homo- and heterodimers with several ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
X-ray Crystallography
X-ray crystallography is the experimental science determining the atomic and molecular structure of a crystal, in which the crystalline structure causes a beam of incident X-rays to diffract into many specific directions. By measuring the angles and intensities of these diffracted beams, a crystallographer can produce a three-dimensional picture of the density of electrons within the crystal. From this electron density, the mean positions of the atoms in the crystal can be determined, as well as their chemical bonds, their crystallographic disorder, and various other information. Since many materials can form crystals—such as salts, metals, minerals, semiconductors, as well as various inorganic, organic, and biological molecules—X-ray crystallography has been fundamental in the development of many scientific fields. In its first decades of use, this method determined the size of atoms, the lengths and types of chemical bonds, and the atomic-scale differences among variou ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Archaea
Archaea ( ; singular archaeon ) is a domain of single-celled organisms. These microorganisms lack cell nuclei and are therefore prokaryotes. Archaea were initially classified as bacteria, receiving the name archaebacteria (in the Archaebacteria kingdom), but this term has fallen out of use. Archaeal cells have unique properties separating them from the other two domains, Bacteria and Eukaryota. Archaea are further divided into multiple recognized phyla. Classification is difficult because most have not been isolated in a laboratory and have been detected only by their gene sequences in environmental samples. Archaea and bacteria are generally similar in size and shape, although a few archaea have very different shapes, such as the flat, square cells of '' Haloquadratum walsbyi''. Despite this morphological similarity to bacteria, archaea possess genes and several metabolic pathways that are more closely related to those of eukaryotes, notably for the enzymes invo ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
C2 Domain
A C2 domain is a protein structural domain involved in targeting proteins to cell membranes. The typical version (PKC-C2) has a beta-sandwich composed of 8 β-strands that co-ordinates two or three calcium ions, which bind in a cavity formed by the first and final loops of the domain, on the membrane binding face. Many other C2 domain families don't have calcium binding activity. Coupling with other domains C2 domains are frequently found coupled to enzymatic domains; for example, the C2 domain in PTEN, brings the phosphatase domain into contact with the plasma membrane, where it can dephosphorylate its substrate, phosphatidylinositol (3,4,5)-trisphosphate (PIP3), without removing it from the membrane - which would be energetically very costly. PTEN consists of two domains, a protein tyrosine phosphatase domain and a C2 domain. This domain pair constitutes a superdomain, a heritable unit that is found in various proteins in fungi, plants and animals. In addition, phosphatid ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Gene Duplication
Gene duplication (or chromosomal duplication or gene amplification) is a major mechanism through which new genetic material is generated during molecular evolution. It can be defined as any duplication of a region of DNA that contains a gene. Gene duplications can arise as products of several types of errors in DNA replication and repair machinery as well as through fortuitous capture by selfish genetic elements. Common sources of gene duplications include ectopic recombination, retrotransposition event, aneuploidy, polyploidy, and replication slippage. Mechanisms of duplication Ectopic recombination Duplications arise from an event termed unequal crossing-over that occurs during meiosis between misaligned homologous chromosomes. The chance of it happening is a function of the degree of sharing of repetitive elements between two chromosomes. The products of this recombination are a duplication at the site of the exchange and a reciprocal deletion. Ectopic recombination ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Transmembrane Domain
A transmembrane domain (TMD) is a membrane-spanning protein domain. TMDs generally adopt an alpha helix topological conformation, although some TMDs such as those in porins can adopt a different conformation. Because the interior of the lipid bilayer is hydrophobic, the amino acid residues in TMDs are often hydrophobic, although proteins such as membrane pumps and ion channels can contain polar residues. TMDs vary greatly in length, sequence, and hydrophobicity, adopting organelle-specific properties. Functions of transmembrane domains Transmembrane domains are known to perform a variety of functions. These include: * Anchoring transmembrane proteins to the membrane. *Facilitating molecular transport of molecules such as ions and proteins across biological membranes; usually hydrophilic residues and binding sites in the TMDs help in this process. *Signal transduction across the membrane; many transmembrane proteins, such as G protein-coupled receptors, receive extracellular ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Hydrophobicity Scales
Hydrophobicity scales are values that define the relative hydrophobicity or hydrophilicity of amino acid residues. The more positive the value, the more hydrophobic are the amino acids located in that region of the protein. These scales are commonly used to predict the transmembrane alpha-helices of membrane proteins. When consecutively measuring amino acids of a protein, changes in value indicate attraction of specific protein regions towards the hydrophobic region inside lipid bilayer. The hydrophobic or hydrophilic character of a compound or amino acid is its hydropathic character, hydropathicity, or hydropathy. Hydrophobicity and the hydrophobic effect The hydrophobic effect represents the tendency of water to exclude non-polar molecules. The effect originates from the disruption of highly dynamic hydrogen bonds between molecules of liquid water. Polar chemical groups, such as OH group in methanol do not cause the hydrophobic effect. However, a pure hydrocarbon molecule, for ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Structure Prediction
Protein structure prediction is the inference of the three-dimensional structure of a protein from its amino acid sequence—that is, the prediction of its secondary and tertiary structure from primary structure. Structure prediction is different from the inverse problem of protein design. Protein structure prediction is one of the most important goals pursued by computational biology; and it is important in medicine (for example, in drug design) and biotechnology (for example, in the design of novel enzymes). Starting in 1994, the performance of current methods is assessed biannually in the CASP experiment (Critical Assessment of Techniques for Protein Structure Prediction). A continuous evaluation of protein structure prediction web servers is performed by the community project CAMEO3D. Protein structure and terminology Proteins are chains of amino acids joined together by peptide bonds. Many conformations of this chain are possible due to the rotation of the main chain ab ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Sodium–potassium Pump
The sodium–potassium pump (sodium–potassium adenosine triphosphatase, also known as Na⁺/K⁺-ATPase, Na⁺/K⁺ pump, or sodium–potassium ATPase) is an enzyme (an electrogenic transmembrane ATPase) found in the membrane of all animal cells. It performs several functions in cell physiology. The Na⁺/K⁺-ATPase enzyme is active (i.e. it uses energy from ATP). For every ATP molecule that the pump uses, three sodium ions are exported and two potassium ions are imported. Thus, there is a net export of a single positive charge per pump cycle. The sodium–potassium pump was discovered in 1957 by the Danish scientist Jens Christian Skou, who was awarded a Nobel Prize for his work in 1997. Its discovery marked an important step forward in the understanding of how ions get into and out of cells, and it has particular significance for excitable cells such as nerve cells, which depend on this pump to respond to stimuli and transmit impulses. All mammals have four different s ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Digoxin
Digoxin (better known as Digitalis), sold under the brand name Lanoxin among others, is a medication used to treat various heart conditions. Most frequently it is used for atrial fibrillation, atrial flutter, and heart failure. Digoxin is one of the oldest medications used in the field of cardiology. It works by increasing myocardial contractility, increasing stroke volume and blood pressure, reducing heart rate, and somewhat extending the time frame of the contraction. Digoxin is taken by mouth or by injection into a vein. Digoxin has a half life of approximately 36 hours given at average doses in patients with normal renal function. It is excreted mostly unchanged in the urine. Common side effects include breast enlargement with other side effects generally due to an excessive dose. These side effects may include loss of appetite, nausea, trouble seeing, confusion, and an irregular heartbeat. Greater care is required in older people and those with poor kidney function. ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |


.jpg)
.png)