|
Single Cell Sequencing
Single-cell sequencing examines the sequence information from individual cells with optimized next-generation sequencing technologies, providing a higher resolution of cellular differences and a better understanding of the function of an individual cell in the context of its microenvironment. For example, in cancer, sequencing the DNA of individual cells can give information about mutations carried by small populations of cells. In development, sequencing the RNAs expressed by individual cells can give insight into the existence and behavior of different cell types. In microbial systems, a population of the same species can appear genetically clonal. Still, single-cell sequencing of RNA or epigenetic modifications can reveal cell-to-cell variability that may help populations rapidly adapt to survive in changing environments. Background A typical human cell consists of about 2 x 3.3 billion base pairs of DNA and 600 million mRNA bases. Usually, a mix of millions of cells is used ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Genome Sequence
In the fields of molecular biology and genetics, a genome is all the genetic information of an organism. It consists of nucleotide sequences of DNA (or RNA in RNA viruses). The nuclear genome includes protein-coding genes and non-coding genes, other functional regions of the genome such as regulatory sequences (see non-coding DNA), and often a substantial fraction of 'junk' DNA with no evident function. Almost all eukaryotes have mitochondria and a small mitochondrial genome. Algae and plants also contain chloroplasts with a chloroplast genome. The study of the genome is called genomics. The genomes of many organisms have been sequenced and various regions have been annotated. The International Human Genome Project reported the sequence of the genome for ''Homo sapiens'' in 200The Human Genome Project although the initial "finished" sequence was missing 8% of the genome consisting mostly of repetitive sequences. With advancements in technology that could handle sequenc ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Polymerases
A polymerase is an enzyme ( EC 2.7.7.6/7/19/48/49) that synthesizes long chains of polymers or nucleic acids. DNA polymerase and RNA polymerase are used to assemble DNA and RNA molecules, respectively, by copying a DNA template strand using base-pairing interactions or RNA by half ladder replication. A DNA polymerase from the thermophilic bacterium, ''Thermus aquaticus'' (''Taq'') ( PDBbr>1BGX EC 2.7.7.7) is used in the polymerase chain reaction, an important technique of molecular biology. A polymerase may be template dependent or template independent. Poly-A-polymerase is an example of template independent polymerase. Terminal deoxynucleotidyl transferase also known to have template independent and template dependent activities. Types By function *DNA polymerase (DNA-directed DNA polymerase, DdDP) **Family A: DNA polymerase I; Pol γ, θ, ν **Family B: DNA polymerase II; Pol α, δ, ε, ζ **Family C: DNA polymerase III holoenzyme **Family X: Pol β, λ, μ ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Bromodeoxyuridine
Bromodeoxyuridine (5-bromo-2'-deoxyuridine, BrdU, BUdR, BrdUrd, broxuridine) is a synthetic nucleoside analogue with a chemical structure similar to thymidine. BrdU is commonly used to study cell proliferation in living tissues and has been studied as a radiosensitizer and diagnostic tool in people with cancer. During S phase of the cell cycle (when DNA replication occurs), BrdU can be incorporated in place of thymidine in newly synthesized DNA molecules of dividing cells. Cells that have recently performed DNA replication or DNA repair can be detected with antibodies specific for BrdU using techniques such as immunohistochemistry or immunofluorescence. BrdU-labelled cells in humans can be detected up to two years after BrdU infusion. Because BrdU can replace thymidine during DNA replication, it can cause mutations, and its use is therefore potentially a health hazard. However, because it is neither radioactive nor myelotoxic at labeling concentrations, it is widely preferre ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Haplotype
A haplotype ( haploid genotype) is a group of alleles in an organism that are inherited together from a single parent. Many organisms contain genetic material ( DNA) which is inherited from two parents. Normally these organisms have their DNA organized in two sets of pairwise similar chromosomes. The offspring gets one chromosome in each pair from each parent. A set of pairs of chromosomes is called diploid and a set of only one half of each pair is called haploid. The haploid genotype (haplotype) is a genotype that considers the singular chromosomes rather than the pairs of chromosomes. It can be all the chromosomes from one of the parents or a minor part of a chromosome, for example a sequence of 9000 base pairs. However, there are other uses of this term. First, it is used to mean a collection of specific alleles (that is, specific DNA sequences) in a cluster of tightly linked genes on a chromosome that are likely to be inherited together—that is, they are likely to be co ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Chromothripsis
Chromothripsis is a mutational process by which up to thousands of clustered chromosomal rearrangements occur in a single event in localised and confined genomic regions in one or a few chromosomes, and is known to be involved in both cancer and congenital diseases. It occurs through one massive genomic rearrangement during a single catastrophic event in the cell's history. It is believed that for the cell to be able to withstand such a destructive event, the occurrence of such an event must be the upper limit of what a cell can tolerate and survive. The chromothripsis phenomenon opposes the conventional theory that cancer is the gradual acquisition of genomic rearrangements and somatic mutations over time. The simplest model as to how these rearrangements occur is through the simultaneous fragmentation of distinct chromosomal regions (breakpoints show a non-random distribution) and then subsequent imperfect reassembly by DNA repair pathways or aberrant DNA replication mechanisms. ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Breakage-fusion-bridge Cycle
Breakage-fusion-bridge (BFB) cycle (also breakage-rejoining-bridge cycle) is a mechanism of chromosomal instability, discovered by Barbara McClintock in the late 1930s. Mechanism The BFB cycle begins when the end region of a chromosome, called its telomere, breaks off. When that chromosome subsequently replicates it forms two sister chromatids which both lack a telomere. Since telomeres appear at the end of chromatids, and function to prevent their ends from fusing with other chromatids, the lack of a telomere on these two sister chromatids causes them to fuse with one another. During anaphase the sister chromatids will form a bridge where the centromere in one of the sister chromatids will be pulled in one direction of the dividing cell, while the centromere of the other will be pulled in the opposite direction. Being pulled in opposite directions will cause the two sister chromatids to break apart from each other, but not necessarily at the site that they fused. This results in ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Allele
An allele (, ; ; modern formation from Greek ἄλλος ''állos'', "other") is a variation of the same sequence of nucleotides at the same place on a long DNA molecule, as described in leading textbooks on genetics and evolution. ::"The chromosomal or genomic location of a gene or any other genetic element is called a locus (plural: loci) and alternative DNA sequences at a locus are called alleles." The simplest alleles are single nucleotide polymorphisms (SNP). but they can also be insertions and deletions of up to several thousand base pairs. Popular definitions of 'allele' typically refer only to different alleles within genes. For example, the ABO blood grouping is controlled by the ABO gene, which has six common alleles (variants). In population genetics, nearly every living human's phenotype for the ABO gene is some combination of just these six alleles. Most alleles observed result in little or no change in the function of the gene product it codes for. However, ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
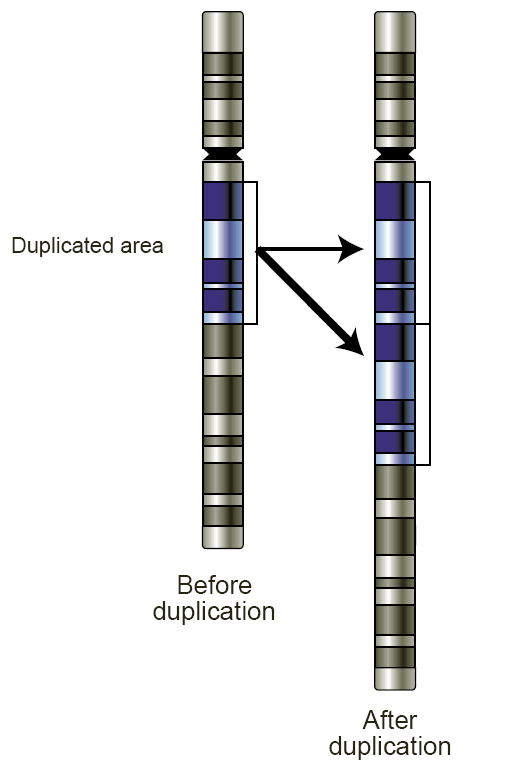
Copy Number Variation
Copy number variation (CNV) is a phenomenon in which sections of the genome are repeated and the number of repeats in the genome varies between individuals. Copy number variation is a type of structural variation: specifically, it is a type of duplication or deletion event that affects a considerable number of base pairs. Approximately two-thirds of the entire human genome may be composed of repeats and 4.8–9.5% of the human genome can be classified as copy number variations. In mammals, copy number variations play an important role in generating necessary variation in the population as well as disease phenotype. Copy number variations can be generally categorized into two main groups: short repeats and long repeats. However, there are no clear boundaries between the two groups and the classification depends on the nature of the loci of interest. Short repeats include mainly dinucleotide repeats (two repeating nucleotides e.g. A-C-A-C-A-C...) and trinucleotide repeats. Long ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Human Genome
The human genome is a complete set of nucleic acid sequences for humans, encoded as DNA within the 23 chromosome pairs in cell nuclei and in a small DNA molecule found within individual mitochondria. These are usually treated separately as the nuclear genome and the mitochondrial genome. Human genomes include both protein-coding DNA sequences and various types of DNA that does not encode proteins. The latter is a diverse category that includes DNA coding for non-translated RNA, such as that for ribosomal RNA, transfer RNA, ribozymes, small nuclear RNAs, and several types of regulatory RNAs. It also includes promoters and their associated gene-regulatory elements, DNA playing structural and replicatory roles, such as scaffolding regions, telomeres, centromeres, and origins of replication, plus large numbers of transposable elements, inserted viral DNA, non-functional pseudogenes and simple, highly-repetitive sequences. Introns make up a large percentage of non-coding DN ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Single-nucleotide Polymorphisms
In genetics, a single-nucleotide polymorphism (SNP ; plural SNPs ) is a germline substitution of a single nucleotide at a specific position in the genome. Although certain definitions require the substitution to be present in a sufficiently large fraction of the population (e.g. 1% or more), many publications do not apply such a frequency threshold. For example, at a specific base position in the human genome, the G nucleotide may appear in most individuals, but in a minority of individuals, the position is occupied by an A. This means that there is a SNP at this specific position, and the two possible nucleotide variations – G or A – are said to be the alleles for this specific position. SNPs pinpoint differences in our susceptibility to a wide range of diseases, for example age-related macular degeneration (a common SNP in the CFH gene is associated with increased risk of the disease) or nonalcoholic fatty liver disease (a SNP in the PNPLA3 gene is associated with incre ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Fluorescent In Situ Hybridization
Fluorescence ''in situ'' hybridization (FISH) is a molecular cytogenetic technique that uses fluorescent probes that bind to only particular parts of a nucleic acid sequence with a high degree of sequence complementarity. It was developed by biomedical researchers in the early 1980s to detect and localize the presence or absence of specific DNA sequences on chromosomes. Fluorescence microscopy can be used to find out where the fluorescent probe is bound to the chromosomes. FISH is often used for finding specific features in DNA for use in genetic counseling, medicine, and species identification. FISH can also be used to detect and localize specific RNA targets ( mRNA, lncRNA and miRNA) in cells, circulating tumor cells, and tissue samples. In this context, it can help define the spatial-temporal patterns of gene expression within cells and tissues. Probes – RNA and DNA In biology, a probe is a single strand of DNA or RNA that is complementary to a nucleotide sequenc ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Single-cell DNA Template Strand Sequencing
Single-cell DNA template strand sequencing, or Strand-seq, is a technique for the selective sequencing of a daughter cell's parental template strands. This technique offers a wide variety of applications, including the identification of sister chromatid exchanges in the parental cell prior to segregation, the assessment of non-random segregation of sister chromatids, the identification of misoriented contigs in genome assemblies, de novo genome assembly of both haplotypes in diploid organisms including humans, whole-chromosome haplotyping, and the identification of germline and somatic genomic structural variation, the latter of which can be detected robustly even in single cells. Background Strand-seq (single-cell and single-strand sequencing) was one of the first single-cell sequencing protocols described in 2012. This genomic technique selectively sequencings the parental template strands in single daughter cells DNA libraries. As a proof of concept study, the authors d ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |