|
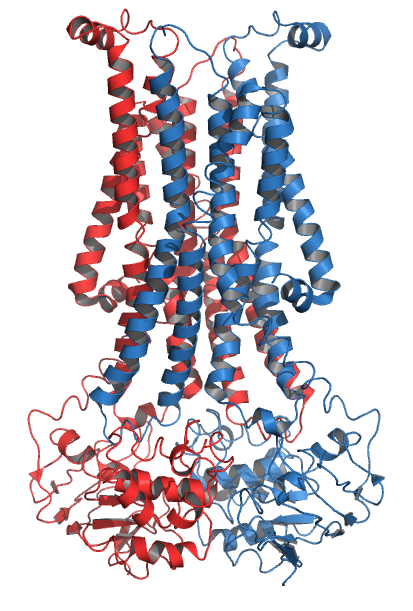
Scramblases
Scramblase is a protein responsible for the translocation of phospholipids between the two monolayers of a lipid bilayer of a cell membrane. In humans, phospholipid scramblases (PLSCRs) constitute a family of five homologous proteins that are named as hPLSCR1–hPLSCR5. Scramblases are not members of the general family of transmembrane lipid transporters known as flippases. Scramblases are distinct from flippases and floppases. Scramblases, flippases, and floppases are three different types of enzymatic groups of phospholipid transportation enzymes. The inner-leaflet, facing the inside of the cell, contains negatively charged amino-phospholipids and phosphatidylethanolamine. The outer-leaflet, facing the outside environment, contains phosphatidylcholine and sphingomyelin. Scramblase is an enzyme, present in the cell membrane, that can transport (''scramble'') the negatively charged phospholipids from the inner-leaflet to the outer-leaflet, and vice versa. Expression Wher ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
PLSCR4
Phospholipid scramblase 4, also known as Ca2+-dependent phospholipid scramblase 4, is a protein that is encoded in humans by the ''PLSCR4'' gene. See also * Scramblase Scramblase is a protein responsible for the translocation of phospholipids between the two monolayers of a lipid bilayer of a cell membrane. In humans, phospholipid scramblases (PLSCRs) constitute a family of five homologous proteins tha ... References Further reading * * * * * * * * * {{gene-3-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
PLSCR2
Phospholipid scramblase 2, also known as Ca2+-dependent phospholipid scramblase 2, is a protein that in humans is encoded by the ''PLSCR2'' gene. See also * scramblase Scramblase is a protein responsible for the translocation of phospholipids between the two monolayers of a lipid bilayer of a cell membrane. In humans, phospholipid scramblases (PLSCRs) constitute a family of five homologous proteins tha ... References Further reading * * * * {{gene-3-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Flippase
Flippases (rarely spelled flipases) are transmembrane lipid transporter proteins located in the membrane which belong to ABC transporter or P4-type ATPase families. They are responsible for aiding the movement of phospholipid molecules between the two leaflets that compose a cell's membrane (transverse diffusion, also known as a "flip-flop" transition). The possibility of active maintenance of an asymmetric distribution of molecules in the phospholipid bilayer was predicted in the early 1970s by Mark Bretscher. Although phospholipids diffuse rapidly in the plane of the membrane, their polar head groups cannot pass easily through the hydrophobic center of the bilayer, limiting their diffusion in this dimension. Some flippases - often instead called scramblases - are energy-independent and bidirectional, causing reversible equilibration of phospholipid between the two sides of the membrane, whereas others are energy-dependent and unidirectional, using energy from ATP hydrolysis to pu ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
PLSCR3
Phospholipid scramblase 3 is an enzyme that in humans is encoded by the ''PLSCR3'' gene (abbreviated to PLS3 in this section). Like the other phospholipid scramblase family members (PLS1, PLS2, PLS4), PLS3 is a type II plasma membrane protein that is rich in proline and integral in apoptosis, or programmed cell death. The regulation of apoptosis is critical for both cell development and tissue homeostasis Although phospholipid scramblase is thought to exist in all eukaryotic cells, PLS3 is a protein that is novel to the mitochondria. This is very important because mitochondria are central in the apoptotic cell pathway. This newly found member of the scramblase family is "responsible for phospholipid translocation between two lipid compartments," the inner mitochondrial membrane and the outer membrane. Further experimental evidence suggests that the mechanism and effectors of PLS3's enzymatic activity are rather nuanced. Effect on mitochondrial cardiolipin Cardiolipin is a mito ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Lipid Bilayer
The lipid bilayer (or phospholipid bilayer) is a thin polar membrane made of two layers of lipid molecules. These membranes are flat sheets that form a continuous barrier around all cells. The cell membranes of almost all organisms and many viruses are made of a lipid bilayer, as are the nuclear membrane surrounding the cell nucleus, and membranes of the membrane-bound organelles in the cell. The lipid bilayer is the barrier that keeps ions, proteins and other molecules where they are needed and prevents them from diffusing into areas where they should not be. Lipid bilayers are ideally suited to this role, even though they are only a few nanometers in width, because they are impermeable to most water-soluble (hydrophilic) molecules. Bilayers are particularly impermeable to ions, which allows cells to regulate salt concentrations and pH by transporting ions across their membranes using proteins called ion pumps. Biological bilayers are usually composed of amphiphilic phosphol ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Tubby Protein
The tubby protein is encoded by the ''TUB'' gene. It is an upstream cell signaling protein common to multicellular eukaryotes. The first ''tubby'' gene was identified in mice, and proteins that are homologous to tubby are known as "tubby-like proteins" (TULPs). They share a common and characteristic tertiary structure that consists of a beta barrel packed around an alpha helix in the central pore. The gene derives its name from its role in metabolism; mice with a mutated tubby gene develop delayed-onset obesity, sensorineural hearing loss and retinal degeneration. Structure Tubby proteins are classified as α+β proteins and have a 12- beta stranded barrel surrounding a central alpha helix. Tubby proteins can bind the small cell signaling molecule phosphatidylinositol, which is typically localized to the cell membrane. A similar structural fold to the Tubby like proteins has been identified in the Scramblase family of proteins. Function Tubby proteins have been implicated as ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Proline
Proline (symbol Pro or P) is an organic acid classed as a proteinogenic amino acid (used in the biosynthesis of proteins), although it does not contain the amino group but is rather a secondary amine. The secondary amine nitrogen is in the protonated form (NH2+) under biological conditions, while the carboxyl group is in the deprotonated −COO− form. The "side chain" from the α carbon connects to the nitrogen forming a pyrrolidine loop, classifying it as a aliphatic amino acid. It is non-essential in humans, meaning the body can synthesize it from the non-essential amino acid L-glutamate. It is encoded by all the codons starting with CC (CCU, CCC, CCA, and CCG). Proline is the only proteinogenic secondary amino acid which is a secondary amine, as the nitrogen atom is attached both to the α-carbon and to a chain of three carbons that together form a five-membered ring. History and etymology Proline was first isolated in 1900 by Richard Willstätter who obtained the amino ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
EF Hand
The EF hand is a helix–loop–helix structural domain or ''motif'' found in a large family of calcium-binding proteins. The EF-hand motif contains a helix–loop–helix topology, much like the spread thumb and forefinger of the human hand, in which the Ca2+ ions are coordinated by ligands within the loop. The motif takes its name from traditional nomenclature used in describing the protein parvalbumin, which contains three such motifs and is probably involved in muscle relaxation via its calcium-binding activity. The EF-hand consists of two alpha helices linked by a short loop region (usually about 12 amino acids) that usually binds calcium ions. EF-hands also appear in each structural domain of the signaling protein calmodulin and in the muscle protein troponin-C. Calcium ion binding site The calcium ion is coordinated in a pentagonal bipyramidal configuration. The six residues involved in the binding are in positions 1, 3, 5, 7, 9 and 12; these residues are denoted by X ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Domain
In molecular biology, a protein domain is a region of a protein's polypeptide chain that is self-stabilizing and that folds independently from the rest. Each domain forms a compact folded three-dimensional structure. Many proteins consist of several domains, and a domain may appear in a variety of different proteins. Molecular evolution uses domains as building blocks and these may be recombined in different arrangements to create proteins with different functions. In general, domains vary in length from between about 50 amino acids up to 250 amino acids in length. The shortest domains, such as zinc fingers, are stabilized by metal ions or disulfide bridges. Domains often form functional units, such as the calcium-binding EF hand domain of calmodulin. Because they are independently stable, domains can be "swapped" by genetic engineering between one protein and another to make chimeric proteins. Background The concept of the domain was first proposed in 1973 by Wetlaufer aft ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Adenosine Triphosphate
Adenosine triphosphate (ATP) is an organic compound that provides energy to drive many processes in living cells, such as muscle contraction, nerve impulse propagation, condensate dissolution, and chemical synthesis. Found in all known forms of life, ATP is often referred to as the "molecular unit of currency" of intracellular energy transfer. When consumed in metabolic processes, it converts either to adenosine diphosphate (ADP) or to adenosine monophosphate (AMP). Other processes regenerate ATP. The human body recycles its own body weight equivalent in ATP each day. It is also a precursor to DNA and RNA, and is used as a coenzyme. From the perspective of biochemistry, ATP is classified as a nucleoside triphosphate, which indicates that it consists of three components: a nitrogenous base (adenine), the sugar ribose, and the Polyphosphate, triphosphate. Structure ATP consists of an adenine attached by the 9-nitrogen atom to the 1′ carbon atom of a sugar (ribose), which i ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
PLSCR1
Phospholipid scramblase 1 (PL scramblase 1) is an enzyme that in humans is encoded by the ''PLSCR1'' gene. Interactions PLSCR1 has been shown to interact with: * CPSF6, * Epidermal growth factor receptor, * NEU4, * SHC1, * SLPI, and * TFG. See also *Scramblase Scramblase is a protein responsible for the translocation of phospholipids between the two monolayers of a lipid bilayer of a cell membrane. In humans, phospholipid scramblases (PLSCRs) constitute a family of five homologous proteins tha ... References Further reading * * * * * * * * * * * * * * * * * * {{gene-3-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Redox
Redox (reduction–oxidation, , ) is a type of chemical reaction in which the oxidation states of substrate (chemistry), substrate change. Oxidation is the loss of Electron, electrons or an increase in the oxidation state, while reduction is the gain of electrons or a decrease in the oxidation state. There are two classes of redox reactions: * ''Electron-transfer'' – Only one (usually) electron flows from the reducing agent to the oxidant. This type of redox reaction is often discussed in terms of redox couples and electrode potentials. * ''Atom transfer'' – An atom transfers from one substrate to another. For example, in the rusting of iron, the oxidation state of iron atoms increases as the iron converts to an oxide, and simultaneously the oxidation state of oxygen decreases as it accepts electrons released by the iron. Although oxidation reactions are commonly associated with the formation of oxides, other chemical species can serve the same function. In hydrogen ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |