|
Protein Disulfide-isomerase
Protein disulfide isomerase (), or PDI, is an enzyme in the endoplasmic reticulum (ER) in eukaryotes and the periplasm of bacteria that catalyzes the formation and breakage of disulfide bonds between cysteine residues within proteins as they fold. This allows proteins to quickly find the correct arrangement of disulfide bonds in their fully folded state, and therefore the enzyme acts to catalyze protein folding. Structure Protein disulfide-isomerase has two catalytic thioredoxin-like domains (active sites), each containing the canonical CGHC motif, and two non catalytic domains. This structure is similar to the structure of enzymes responsible for oxidative folding in the intermembrane space of the mitochondria; an example of this is mitochondrial IMS import and assembly (Mia40), which has 2 catalytic domains that contain a CX9C, which is similar to the CGHC domain of PDI. Bacterial DsbA, responsible for oxidative folding, also has a thioredoxin CXXC domain. Function ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
PDIA2
Protein disulfide isomerase family A member 2 is a protein that in humans is encoded by the PDIA2 gene. Function This gene encodes a member of the Protein disulfide-isomerase, disulfide isomerase (PDI) family of endoplasmic reticulum (ER) proteins that catalyze protein folding and thiol-disulfide interchange reactions. The encoded protein has an N-terminal ER-signal sequence, two catalytically active thioredoxin (TRX) domains, two TRX-like domains and a C-terminal ER-retention sequence. The protein plays a role in the folding of nascent proteins in the endoplasmic reticulum by forming disulfide bonds through its thiol isomerase, oxidase, and reductase activity. The encoded protein also possesses estradiol-binding activity and can modulate intracellular estradiol levels. [provided by RefSeq, Sep 2017]. References Further reading * * * * * * * * * {{gene-16-stub Endoplasmic reticulum resident proteins ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Oxidoreductase
In biochemistry, an oxidoreductase is an enzyme that catalyzes the transfer of electrons from one molecule, the reductant, also called the electron donor, to another, the oxidant, also called the electron acceptor. This group of enzymes usually utilizes NADP+ or NAD+ as cofactors. Transmembrane oxidoreductases create electron transport chains in bacteria, chloroplasts and mitochondria, including respiratory complexes I, II and III. Some others can associate with biological membranes as peripheral membrane proteins or be anchored to the membranes through a single transmembrane helix. in ...
|
Antigenic Peptide
In immunology, an antigen (Ag) is a molecule or molecular structure or any foreign particulate matter or a pollen grain that can bind to a specific antibody or T-cell receptor. The presence of antigens in the body may trigger an immune response. The term ''antigen'' originally referred to a substance that is an antibody generator. Antigens can be proteins, peptides (amino acid chains), polysaccharides (chains of monosaccharides/simple sugars), lipids, or nucleic acids. Antigens are recognized by antigen receptors, including antibodies and T-cell receptors. Diverse antigen receptors are made by cells of the immune system so that each cell has a specificity for a single antigen. Upon exposure to an antigen, only the lymphocytes that recognize that antigen are activated and expanded, a process known as clonal selection. In most cases, an antibody can only react to and bind one specific antigen; in some instances, however, antibodies may cross-react and bind more than one antigen. T ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
D1 Protein
D1, D01, D.I, D.1 or D-1 can refer to: Science and technology Biochemistry and medicine * ATC code D01 ''Antifungals for dermatological use'', a subgroup of the Anatomical Therapeutic Chemical Classification System * Dopamine receptor D1, a protein * Haplogroup D1 (Y-DNA) * Vitamin D1, a form of Vitamin D * DI, Iodothyronine deiodinase type I, an enzyme involved with thyroid hormones Technology * Nikon D1, a digital single-lens reflex camera * D1, former brand of T-Mobile in Germany * D1, an abbreviation for DOCSIS 1.0 1.0, an international telecommunications standard * D-1 (Sony), an early digital video recording format * STS-61-A, also known as D-1, the 22nd mission of NASA's Space Shuttle program * D-1, from the Proton (rocket family), Russian rockets * Mercedes D.I, a 1913 German aircraft engine Military World War I fighter aircraft * AEG D.I * Albatros D.I * Halberstadt D.I, experimental version of Halberstadt D.II (and Aviatik D.I variant) * Aviatik (Berg) D.I * Daimler ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
RNA-binding Protein
RNA-binding proteins (often abbreviated as RBPs) are proteins that bind to the double or single stranded RNA in cells and participate in forming ribonucleoprotein complexes. RBPs contain various structural motifs, such as RNA recognition motif (RRM), dsRNA binding domain, zinc finger and others. They are cytoplasmic and nuclear proteins. However, since most mature RNA is exported from the nucleus relatively quickly, most RBPs in the nucleus exist as complexes of protein and pre-mRNA called heterogeneous ribonucleoprotein particles (hnRNPs). RBPs have crucial roles in various cellular processes such as: cellular function, transport and localization. They especially play a major role in post-transcriptional control of RNAs, such as: splicing, polyadenylation, mRNA stabilization, mRNA localization and translation. Eukaryotic cells express diverse RBPs with unique RNA-binding activity and protein–protein interaction. According to the Eukaryotic RBP Database (EuRBPDB), there are 2 ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
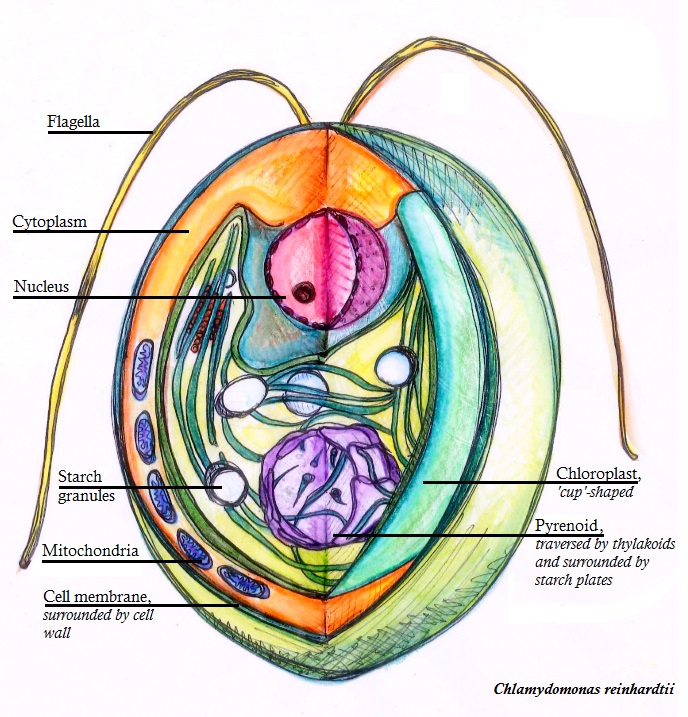
Chlamydomonas Reinhardtii
''Chlamydomonas reinhardtii'' is a single-cell green alga about 10 micrometres in diameter that swims with two flagella. It has a cell wall made of hydroxyproline-rich glycoproteins, a large cup-shaped chloroplast, a large pyrenoid, and an eyespot that senses light. '' Chlamydomonas'' species are widely distributed worldwide in soil and fresh water. ''Chlamydomonas reinhardtii'' is an especially well studied biological model organism, partly due to its ease of culturing and the ability to manipulate its genetics. When illuminated, ''C. reinhardtii'' can grow photoautotrophically, but it can also grow in the dark if supplied with organic carbon. Commercially, ''C. reinhardtii'' is of interest for producing biopharmaceuticals and biofuel, as well being a valuable research tool in making hydrogen. History The ''C. reinhardtii'' wild-type laboratory strain c137 (mt+) originates from an isolate collected near Amherst, Massachusetts, in 1945 by Gilbert M. Smith. The species' n ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Algae
Algae (; singular alga ) is an informal term for a large and diverse group of photosynthetic eukaryotic organisms. It is a polyphyletic grouping that includes species from multiple distinct clades. Included organisms range from unicellular microalgae, such as ''Chlorella,'' ''Prototheca'' and the diatoms, to multicellular forms, such as the giant kelp, a large brown alga which may grow up to in length. Most are aquatic and autotrophic (they generate food internally) and lack many of the distinct cell and tissue types, such as stomata, xylem and phloem that are found in land plants. The largest and most complex marine algae are called seaweeds, while the most complex freshwater forms are the ''Charophyta'', a division of green algae which includes, for example, ''Spirogyra'' and stoneworts. No definition of algae is generally accepted. One definition is that algae "have chlorophyll ''a'' as their primary photosynthetic pigment and lack a sterile covering of cells around thei ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Chloroplast
A chloroplast () is a type of membrane-bound organelle known as a plastid that conducts photosynthesis mostly in plant and algal cells. The photosynthetic pigment chlorophyll captures the energy from sunlight, converts it, and stores it in the energy-storage molecules ATP and NADPH while freeing oxygen from water in the cells. The ATP and NADPH is then used to make organic molecules from carbon dioxide in a process known as the Calvin cycle. Chloroplasts carry out a number of other functions, including fatty acid synthesis, amino acid synthesis, and the immune response in plants. The number of chloroplasts per cell varies from one, in unicellular algae, up to 100 in plants like ''Arabidopsis'' and wheat. A chloroplast is characterized by its two membranes and a high concentration of chlorophyll. Other plastid types, such as the leucoplast and the chromoplast, contain little chlorophyll and do not carry out photosynthesis. Chloroplasts are highly dynamic—they circulat ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Disulfide Bond
In biochemistry, a disulfide (or disulphide in British English) refers to a functional group with the structure . The linkage is also called an SS-bond or sometimes a disulfide bridge and is usually derived by the coupling of two thiol groups. In biology, disulfide bridges formed between thiol groups in two cysteine residues are an important component of the secondary and tertiary structure of proteins. ''Persulfide'' usually refers to compounds. In inorganic chemistry disulfide usually refers to the corresponding anion (−S−S−). Organic disulfides Symmetrical disulfides are compounds of the formula . Most disulfides encountered in organo sulfur chemistry are symmetrical disulfides. Unsymmetrical disulfides (also called heterodisulfides) are compounds of the formula . They are less common in organic chemistry, but most disulfides in nature are unsymmetrical. Properties The disulfide bonds are strong, with a typical bond dissociation energy of 60 kcal/mol (251& ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Post-translational Modification
Post-translational modification (PTM) is the covalent and generally enzymatic modification of proteins following protein biosynthesis. This process occurs in the endoplasmic reticulum and the golgi apparatus. Proteins are synthesized by ribosomes translating mRNA into polypeptide chains, which may then undergo PTM to form the mature protein product. PTMs are important components in cell signaling, as for example when prohormones are converted to hormones. Post-translational modifications can occur on the amino acid side chains or at the protein's C- or N- termini. They can extend the chemical repertoire of the 20 standard amino acids by modifying an existing functional group or introducing a new one such as phosphate. Phosphorylation is a highly effective mechanism for regulating the activity of enzymes and is the most common post-translational modification. Many eukaryotic and prokaryotic proteins also have carbohydrate molecules attached to them in a process called glycosyla ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
N Terminus
The N-terminus (also known as the amino-terminus, NH2-terminus, N-terminal end or amine-terminus) is the start of a protein or polypeptide, referring to the free amine group (-NH2) located at the end of a polypeptide. Within a peptide, the amine group is bonded to the carboxylic group of another amino acid, making it a chain. That leaves a free carboxylic group at one end of the peptide, called the C-terminus, and a free amine group on the other end called the N-terminus. By convention, peptide sequences are written N-terminus to C-terminus, left to right (in LTR writing systems). This correlates the translation direction to the text direction, because when a protein is translated from messenger RNA, it is created from the N-terminus to the C-terminus, as amino acids are added to the carboxyl end of the protein. Chemistry Each amino acid has an amine group and a carboxylic group. Amino acids link to one another by peptide bonds which form through a dehydration reaction that jo ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |