|
Nuclear Organization
Nuclear organization refers to the spatial distribution of chromatin within a cell nucleus. There are many different levels and scales of nuclear organisation. Chromatin is a higher order structure of DNA. At the smallest scale, DNA is packaged into units called nucleosomes. The quantity and organisation of these nucleosomes can affect the accessibility of local chromatin. This has a knock-on effect on the expression of nearby genes, additionally determining whether or not they can be regulated by transcription factors. At slightly larger scales, DNA looping can physically bring together DNA elements that would otherwise be separated by large distances. These interactions allow regulatory signals to cross over large genomic distances—for example, from enhancers to promoters. In contrast, on a large scale, the arrangement of chromosomes can determine their properties. Chromosomes are organised into two compartments labelled A ("active") and B ("inactive"), each with dis ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Nuclear Technology
Nuclear technology is technology that involves the nuclear reactions of atomic nuclei. Among the notable nuclear technologies are nuclear reactors, nuclear medicine and nuclear weapons. It is also used, among other things, in smoke detectors and gun sights. History and scientific background Discovery The vast majority of common, natural phenomena on Earth only involve gravity and electromagnetism, and not nuclear reactions. This is because atomic nuclei are generally kept apart because they contain positive electrical charges and therefore repel each other. In 1896, Henri Becquerel was investigating phosphorescence in uranium salts when he discovered a new phenomenon which came to be called radioactivity. He, Pierre Curie and Marie Curie began investigating the phenomenon. In the process, they isolated the element radium, which is highly radioactive. They discovered that radioactive materials produce intense, penetrating rays of three distinct sorts, which they labeled al ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Nucleic Acid Sequence
A nucleic acid sequence is a succession of Nucleobase, bases signified by a series of a set of five different letters that indicate the order of nucleotides forming alleles within a DNA (using GACT) or RNA (GACU) molecule. By convention, sequences are usually presented from the Directionality (molecular biology), 5' end to the 3' end. For DNA, the Sense (molecular biology), sense strand is used. Because nucleic acids are normally linear (unbranched) polymers, specifying the sequence is equivalent to defining the covalent structure of the entire molecule. For this reason, the nucleic acid sequence is also termed the Biomolecular structure#Primary structure, primary structure. The sequence has capacity to represent information. Biological deoxyribonucleic acid represents the information which directs the functions of an organism. Nucleic acids also have a Nucleic acid secondary structure, secondary structure and Nucleic acid tertiary structure, tertiary structure. Primary structur ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
CRISPR
CRISPR () (an acronym for clustered regularly interspaced short palindromic repeats) is a family of DNA sequences found in the genomes of prokaryotic organisms such as bacteria and archaea. These sequences are derived from DNA fragments of bacteriophages that had previously infected the prokaryote. They are used to detect and destroy DNA from similar bacteriophages during subsequent infections. Hence these sequences play a key role in the antiviral (i.e. anti-phage) defense system of prokaryotes and provide a form of acquired immunity. CRISPR is found in approximately 50% of sequenced bacterial genomes and nearly 90% of sequenced archaea. Cas9 (or "CRISPR-associated protein 9") is an enzyme that uses CRISPR sequences as a guide to recognize and cleave specific strands of DNA that are complementary to the CRISPR sequence. Cas9 enzymes together with CRISPR sequences form the basis of a technology known as CRISPR-Cas9 that can be used to edit genes within organisms. This editing ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Genome Editing
Genome editing, or genome engineering, or gene editing, is a type of genetic engineering in which DNA is inserted, deleted, modified or replaced in the genome of a living organism. Unlike early genetic engineering techniques that randomly inserts genetic material into a host genome, genome editing targets the insertions to site-specific locations. The basic mechanism involved in genetic manipulations through programmable nucleases is the recognition of target genomic loci and binding of effector DNA-binding domain (DBD), double-strand breaks (DSBs) in target DNA by the restriction endonucleases ( FokI and Cas), and the repair of DSBs through homology-directed recombination (HDR) or non-homologous end joining (NHEJ). History Genome editing was pioneered in the 1990s, before the advent of the common current nuclease-based gene editing platforms, however, its use was limited by low efficiencies of editing. Genome editing with engineered nucleases, i.e. all three major classes of ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Chromosome Conformation Capture
Chromosome conformation capture techniques (often abbreviated to 3C technologies or 3C-based methods) are a set of molecular biology methods used to analyze the spatial organization of chromatin in a cell. These methods quantify the number of interactions between genomic loci that are nearby in 3-D space, but may be separated by many nucleotides in the linear genome. Such interactions may result from biological functions, such as promoter- enhancer interactions, or from random polymer looping, where undirected physical motion of chromatin causes loci to collide. Interaction frequencies may be analyzed directly, or they may be converted to distances and used to reconstruct 3-D structures. The chief difference between 3C-based methods is their scope. For example, when using PCR to detect interaction in a 3C experiment, the interactions between two specific fragments are quantified. In contrast, Hi-C quantifies interactions between all possible pairs of fragments simultaneously. ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
High-throughput Sequencing
DNA sequencing is the process of determining the nucleic acid sequence – the order of nucleotides in DNA. It includes any method or technology that is used to determine the order of the four bases: adenine, guanine, cytosine, and thymine. The advent of rapid DNA sequencing methods has greatly accelerated biological and medical research and discovery. Knowledge of DNA sequences has become indispensable for basic biological research, DNA Genographic Projects and in numerous applied fields such as medical diagnosis, biotechnology, forensic biology, virology and biological systematics. Comparing healthy and mutated DNA sequences can diagnose different diseases including various cancers, characterize antibody repertoire, and can be used to guide patient treatment. Having a quick way to sequence DNA allows for faster and more individualized medical care to be administered, and for more organisms to be identified and cataloged. The rapid speed of sequencing attained with modern DNA ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Fluorescence In Situ Hybridization
Fluorescence ''in situ'' hybridization (FISH) is a molecular cytogenetic technique that uses fluorescent probes that bind to only particular parts of a nucleic acid sequence with a high degree of sequence complementarity. It was developed by biomedical researchers in the early 1980s to detect and localize the presence or absence of specific DNA sequences on chromosomes. Fluorescence microscopy can be used to find out where the fluorescent probe is bound to the chromosomes. FISH is often used for finding specific features in DNA for use in genetic counseling, medicine, and species identification. FISH can also be used to detect and localize specific RNA targets (mRNA, lncRNA and miRNA) in cells, circulating tumor cells, and tissue samples. In this context, it can help define the spatial-temporal patterns of gene expression within cells and tissues. Probes – RNA and DNA In biology, a probe is a single strand of DNA or RNA that is complementary to a nucleotide sequence o ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Theodor Boveri
Theodor Heinrich Boveri (12 October 1862 – 15 October 1915) was a German zoologist, comparative anatomist and co-founder of modern cytology. He was notable for the first hypothesis regarding cellular processes that cause cancer, and for describing ''chromatin diminution in nematodes''. Boveri was married to the American biologist Marcella O'Grady (1863–1950). Their daughter Margret Boveri (1900–1975) became one of the best-known journalists in post-World War II Germany. Work Using an optical microscope, Boveri examined the processes involved in the fertilization of the animal egg cell; his favorite research objects were the nematode ''Parascaris'' and sea urchins. Boveri's work with sea urchins showed that it was necessary to have all chromosomes present in order for proper embryonic development to take place. This discovery was an important part of the Boveri–Sutton chromosome theory. He also discovered, in 1888, the importance of the centrosome for the formation of th ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Carl Rabl
Carl Rabl (2 May 1853 in Wels, Austria – 24 December 1917 in Leipzig, Germany Carl Rabl at ) was an Austrian anatomist. His most notable achievement was on the structural consistency of s during the . In 1885 he published that chromosomes do not lose their identity, even though they are no longer visible through the microscope. As a student, Rabl's influences included ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
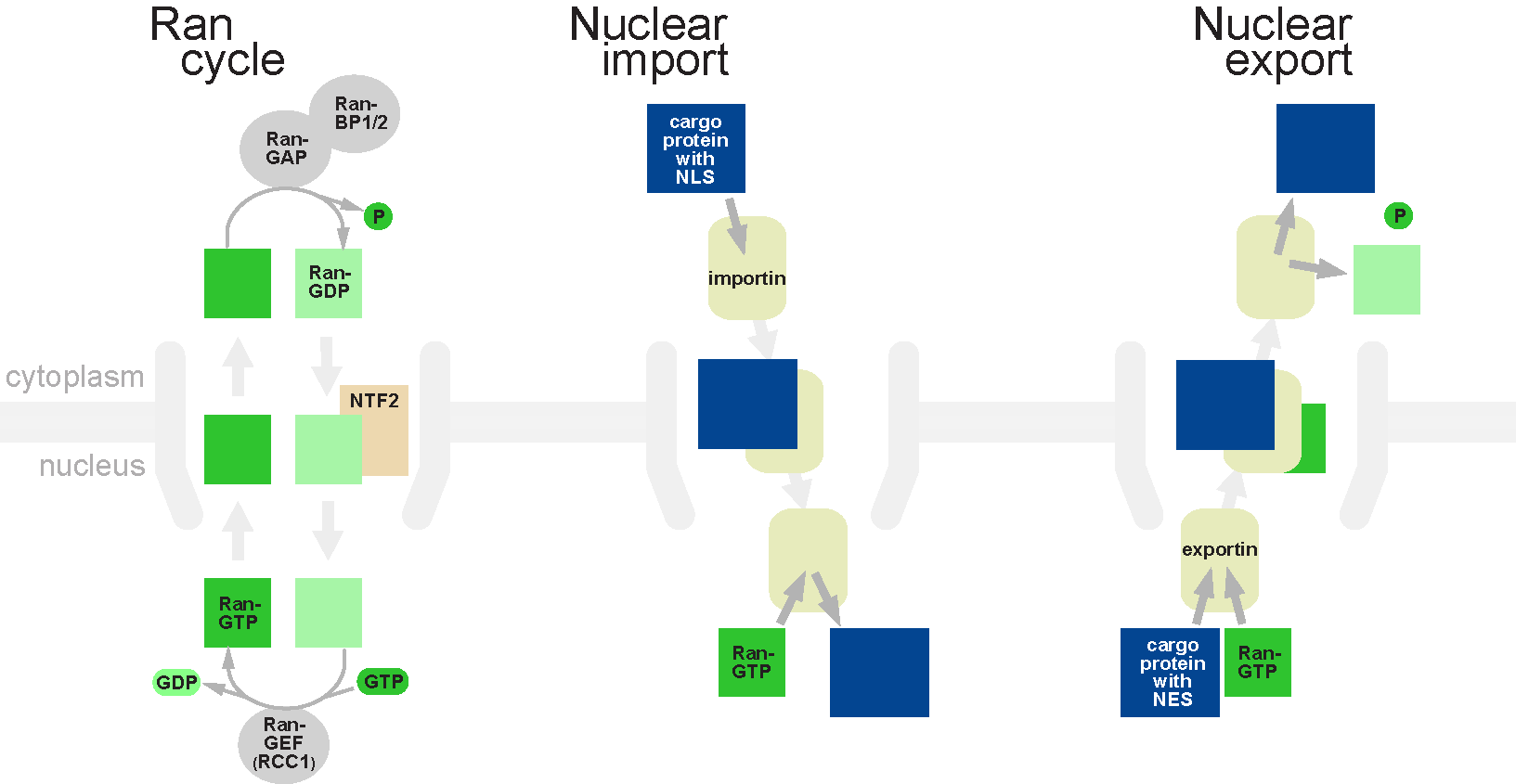
Nuclear Transport
Nuclear transport refers to the mechanisms by which molecules move across the nuclear membrane of a cell. The entry and exit of large molecules from the cell nucleus is tightly controlled by the nuclear pore complexes (NPCs). Although small molecules can enter the nucleus without regulation, macromolecules such as RNA and proteins require association with transport factors known as nuclear transport receptors, like karyopherins called importins to enter the nucleus and exportins to exit. Nuclear import Protein that must be imported to the nucleus from the cytoplasm carry nuclear localization signals (NLS) that are bound by importins. An NLS is a sequence of amino acids that acts as a tag. They are most commonly hydrophilic proteins containing lysine and arginine residues, although diverse NLS sequences have been documented. Proteins, transfer RNA, and assembled ribosomal subunits are exported from the nucleus due to association with exportins, which bind signaling sequences called ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DNA Replication
In molecular biology, DNA replication is the biological process of producing two identical replicas of DNA from one original DNA molecule. DNA replication occurs in all living organisms acting as the most essential part for biological inheritance. This is essential for cell division during growth and repair of damaged tissues, while it also ensures that each of the new cells receives its own copy of the DNA. The cell possesses the distinctive property of division, which makes replication of DNA essential. DNA is made up of a double helix of two complementary strands. The double helix describes the appearance of a double-stranded DNA which is thus composed of two linear strands that run opposite to each other and twist together to form. During replication, these strands are separated. Each strand of the original DNA molecule then serves as a template for the production of its counterpart, a process referred to as semiconservative replication. As a result of semi-conservative rep ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cell Cycle
The cell cycle, or cell-division cycle, is the series of events that take place in a cell that cause it to divide into two daughter cells. These events include the duplication of its DNA (DNA replication) and some of its organelles, and subsequently the partitioning of its cytoplasm, chromosomes and other components into two daughter cells in a process called cell division. In cells with nuclei ( eukaryotes, i.e., animal, plant, fungal, and protist cells), the cell cycle is divided into two main stages: interphase and the mitotic (M) phase (including mitosis and cytokinesis). During interphase, the cell grows, accumulating nutrients needed for mitosis, and replicates its DNA and some of its organelles. During the mitotic phase, the replicated chromosomes, organelles, and cytoplasm separate into two new daughter cells. To ensure the proper replication of cellular components and division, there are control mechanisms known as cell cycle checkpoints after each of the key steps ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |




.png)