|
Mitosome
A mitosome is an organelle found in some unicellular eukaryotic organisms, like in members of the supergroup Excavata. The mitosome was found and named in 1999, and its function has not yet been well characterized. It was termed a ''crypton'' by one group, but that name is no longer in use. The mitosome has been detected only in anaerobic or microaerophilic organisms that do not have mitochondria. These organisms do not have the capability of gaining energy from oxidative phosphorylation, which is normally performed by mitochondria. The mitosome was first described in ''Entamoeba histolytica,'' an intestinal parasite of humans. Mitosomes have also been identified in several species of Microsporidia and in ''Giardia intestinalis''. Origin and function Mitosomes are almost certainly derived from mitochondria. Like mitochondria, they have a double membrane and most proteins are delivered to them by a targeting sequence of amino acids. The targeting sequence is similar to that used ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
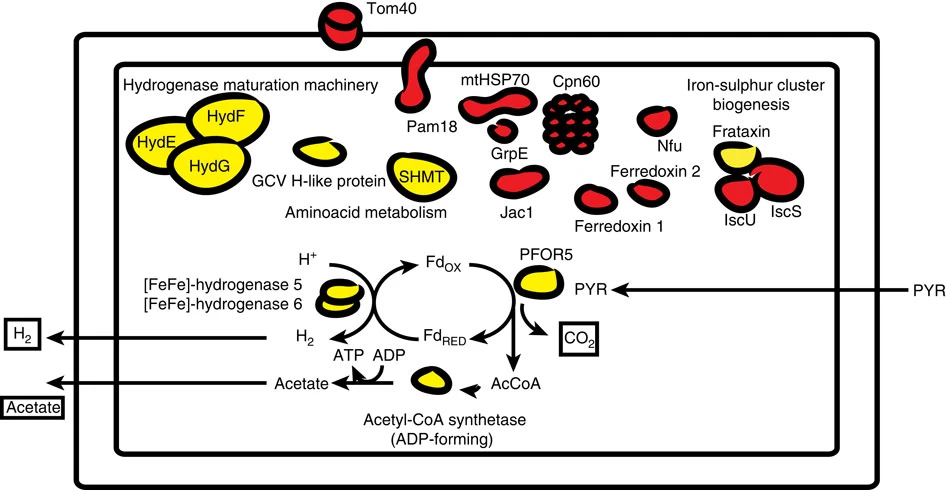
Hydrogenosome
A hydrogenosome is a membrane-enclosed organelle found in some anaerobic ciliates, flagellates, and fungi. Hydrogenosomes are highly variable organelles that have presumably evolved from protomitochondria to produce molecular hydrogen and ATP in anaerobic conditions. Hydrogenosomes were discovered in 1973 by D. G. Lindmark and M. Müller. Because hydrogenosomes hold evolutionary lineage significance for organisms living in anaerobic or oxygen-stressed environments, many research institutions have since documented their findings on how the organelle differs in various sources. History Hydrogenosomes were isolated, purified, biochemically characterized and named in the early 1970s by Lindmark and Müller at Rockefeller University. In addition to this seminal study on hydrogenosomes, they also demonstrated for the first time the presence of pyruvate:ferredoxin oxido-reductase and hydrogenase in eukaryotes. Further studies were subsequently conducted on the biochemical cytology a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Iron-sulfur Protein
Iron–sulfur proteins (or iron–sulphur proteins in British spelling) are proteins characterized by the presence of iron–sulfur clusters containing sulfide-linked di-, tri-, and tetrairon centers in variable oxidation states. Iron–sulfur clusters are found in a variety of metalloproteins, such as the ferredoxins, as well as NADH dehydrogenase, hydrogenases, coenzyme Q – cytochrome c reductase, succinate – coenzyme Q reductase and nitrogenase. Iron–sulfur clusters are best known for their role in the oxidation-reduction reactions of electron transport in mitochondria and chloroplasts. Both Complex I and Complex II of oxidative phosphorylation have multiple Fe–S clusters. They have many other functions including catalysis as illustrated by aconitase, generation of radicals as illustrated by SAM-dependent enzymes, and as sulfur donors in the biosynthesis of lipoic acid and biotin. Additionally, some Fe–S proteins regulate gene expression. Fe–S proteins are vulnerabl ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Organelle
In cell biology, an organelle is a specialized subunit, usually within a cell, that has a specific function. The name ''organelle'' comes from the idea that these structures are parts of cells, as organs are to the body, hence ''organelle,'' the suffix ''-elle'' being a diminutive. Organelles are either separately enclosed within their own lipid bilayers (also called membrane-bound organelles) or are spatially distinct functional units without a surrounding lipid bilayer (non-membrane bound organelles). Although most organelles are functional units within cells, some function units that extend outside of cells are often termed organelles, such as cilia, the flagellum and archaellum, and the trichocyst. Organelles are identified by microscopy, and can also be purified by cell fractionation. There are many types of organelles, particularly in eukaryotic cells. They include structures that make up the endomembrane system (such as the nuclear envelope, endoplasmic reticulum, and G ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Hsp70
The 70 kilodalton heat shock proteins (Hsp70s or DnaK) are a family of conserved ubiquitously expressed heat shock proteins. Proteins with similar structure exist in virtually all living organisms. Intracellularly localized Hsp70s are an important part of the cell's machinery for protein folding, performing chaperoning functions, and helping to protect cells from the adverse effects of physiological stresses. Additionally, membrane-bound Hsp70s have been identified as a potential target for cancer therapies and their extracellularly localized counterparts have been identified as having both membrane-bound and membrane-free structures. Discovery Members of the Hsp70 family are very strongly upregulated by heat stress and toxic chemicals, particularly heavy metals such as arsenic, cadmium, copper, mercury, etc. Heat shock was originally discovered by Ferruccio Ritossa in the 1960s when a lab worker accidentally boosted the incubation temperature of Drosophila (fruit flies). When ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Isu1
Iron-sulfur cluster assembly enzyme ISCU, mitochondrial is a protein that in humans is encoded by the ''ISCU'' gene. It encodes an iron-sulfur (Fe-S) cluster scaffold protein involved in 2Fe-2S_cluster">2Fe-2S.html" ;"title="2Fe-2S_cluster.html" ;"title="nowiki/>2Fe-2S cluster">2Fe-2S">2Fe-2S_cluster.html" ;"title="nowiki/>2Fe-2S cluster">2Fe-2Sand [4Fe-4S] cluster synthesis and maturation. A deficiency of ISCU is associated with a mitochondrial myopathy with lifelong exercise intolerance where only minor exertion causes tachycardia, shortness of breath, muscle weakness and myalgia. Updated 2011 Sep 1 Structure ''ISCU'' is located on the q arm of chromosome 12 in position 23.3 and has 8 exons. ISCU, the protein encoded by this gene, is a member of the NifU family. It is an iron-sulfur transferase that contains binding sites for 2Fe-2S_cluster">2Fe-2S.html" ;"title="2Fe-2S_cluster.html" ;"title="nowiki/>2Fe-2S cluster">2Fe-2S">2Fe-2S_cluster.html" ;"title="nowiki/>2Fe-2S clus ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cysteine Desulfurase
Cysteine (symbol Cys or C; ) is a semiessential proteinogenic amino acid with the formula . The thiol side chain in cysteine often participates in enzymatic reactions as a nucleophile. When present as a deprotonated catalytic residue, sometimes the symbol Cyz is used. The deprotonated form can generally be described by the symbol Cym as well. The thiol is susceptible to oxidation to give the disulfide derivative cystine, which serves an important structural role in many proteins. In this case, the symbol Cyx is sometimes used. When used as a food additive, it has the E number E920. Cysteine is encoded by the codons UGU and UGC. The sulfur-containing amino acids cysteine and methionine are more easily oxidized than the other amino acids. Structure Like other amino acids (not as a residue of a protein), cysteine exists as a zwitterion. Cysteine has chirality in the older / notation based on homology to - and -glyceraldehyde. In the newer ''R''/''S'' system of designating chir ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Frataxin
Frataxin is a protein that in humans is encoded by the FXN gene. It is located in the mitochondrion and Frataxin mRNA is mostly expressed in tissues with a high metabolic rate. The function of frataxin is not clear but it is involved in assembly of iron-sulfur clusters. It has been proposed to act as either an iron chaperone or an iron storage protein. Reduced expression of frataxin is the cause of Friedreich's ataxia. Structure X-ray crystallography has shown that human frataxin consists of a β-sheet that supports a pair of parallel α-helices, forming a compact αβ sandwich. Frataxin sequence homology, homologues in other species are similar, sharing the same core structure. However, the frataxin tail sequences, extending from the end of one helix, diverge in sequence and differ in length. Human frataxin has a longer tail sequence than frataxin found in bacteria or yeast. It is hypothesized that the purpose of the tail is to stabilize the protein. Like most mitocho ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Iron-sulfur Cluster Biosynthesis Protein Family
In biochemistry, the iron-sulfur cluster biosynthesis describes the components and processes involved in the biosynthesis of iron-sulfur proteins. The topic is of interest because these proteins are pervasive. The iron sulfur proteins contain iron-sulfur clusters, some with elaborate structures, that feature iron and sulfide centers. One broad biosynthetic task is producing sulfide (S2-), which requires various families of enzymes. Another broad task is affixing the sulfide to iron, which is achieved on scaffolds, which are nonfunctional. Finally these Fe-S cluster is transferred to a target protein, which then become functional. The formation of iron-sulfur clusters are produced by one of four pathways: *Nitrogen fixation (NIF) system, which is also found in bacteria that are not nitrogen-fixing. *Iron-sulfur cluster (ISC) system, in bacterial and mitochondria *Sulfur assimilation (SUF) system, in plastids and some bacteria In addition to those three systems, the so-called Cy ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Haem Biosynthesis
Heme, or haem (pronounced /Help:IPA/English, hi:m/ ), is a precursor (chemistry), precursor to hemoglobin, which is necessary to bind oxygen in the bloodstream. Heme is biosynthesized in both the bone marrow and the liver. In biochemical terms, heme is a coordination complex "consisting of an iron ion coordinated to a porphyrin acting as a tetradentate ligand, and to one or two axial ligands." The definition is loose, and many depictions omit the axial ligands. Among the metalloporphyrins deployed by metalloproteins as prosthetic groups, heme is one of the most widely used and defines a family of proteins known as hemoproteins. Hemes are most commonly recognized as components of hemoglobin, the red pigment in blood, but are also found in a number of other biologically important hemoproteins such as myoglobin, cytochromes, catalases, heme peroxidase, and Endothelial NOS, endothelial nitric oxide synthase. The word ''haem'' is derived from Ancient Greek language, Greek ''haima'' ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Aerobic Respiration
Cellular respiration is the process by which biological fuels are oxidised in the presence of an inorganic electron acceptor such as oxygen to produce large amounts of energy, to drive the bulk production of ATP. Cellular respiration may be described as a set of metabolic reactions and processes that take place in the cells of organisms to convert chemical energy from nutrients into adenosine triphosphate (ATP), and then release waste products. The reactions involved in respiration are catabolic reactions, which break large molecules into smaller ones, releasing energy. Respiration is one of the key ways a cell releases chemical energy to fuel cellular activity. The overall reaction occurs in a series of biochemical steps, some of which are redox reactions. Although cellular respiration is technically a combustion reaction, it is an unusual one because of the slow, controlled release of energy from the series of reactions. Nutrients that are commonly used by animal and pla ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Signal Peptide
A signal peptide (sometimes referred to as signal sequence, targeting signal, localization signal, localization sequence, transit peptide, leader sequence or leader peptide) is a short peptide (usually 16-30 amino acids long) present at the N-terminus (or occasionally nonclassically at the C-terminus or internally) of most newly synthesized proteins that are destined toward the secretory pathway. These proteins include those that reside either inside certain organelles (the endoplasmic reticulum, Golgi or endosomes), secreted from the cell, or inserted into most cellular membranes. Although most type I membrane-bound proteins have signal peptides, the majority of type II and multi-spanning membrane-bound proteins are targeted to the secretory pathway by their first transmembrane domain, which biochemically resembles a signal sequence except that it is not cleaved. They are a kind of target peptide. Function (translocation) Signal peptides function to prompt a cell to translo ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Eukaryote
Eukaryotes () are organisms whose cells have a nucleus. All animals, plants, fungi, and many unicellular organisms, are Eukaryotes. They belong to the group of organisms Eukaryota or Eukarya, which is one of the three domains of life. Bacteria and Archaea (both prokaryotes) make up the other two domains. The eukaryotes are usually now regarded as having emerged in the Archaea or as a sister of the Asgard archaea. This implies that there are only two domains of life, Bacteria and Archaea, with eukaryotes incorporated among archaea. Eukaryotes represent a small minority of the number of organisms, but, due to their generally much larger size, their collective global biomass is estimated to be about equal to that of prokaryotes. Eukaryotes emerged approximately 2.3–1.8 billion years ago, during the Proterozoic eon, likely as flagellated phagotrophs. Their name comes from the Greek εὖ (''eu'', "well" or "good") and κάρυον (''karyon'', "nut" or "kernel"). Euka ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |