|
Mediator Complex
Mediator is a multiprotein complex that functions as a transcriptional coactivator in all eukaryotes. It was discovered in 1990 in the lab of Roger D. Kornberg, recipient of the 2006 Nobel Prize in Chemistry. Mediator complexes interact with transcription factors and RNA polymerase II. The main function of mediator complexes is to transmit signals from the transcription factors to the polymerase. Mediator complexes are variable at the evolutionary, compositional and conformational levels. The first image shows only one "snapshot" of what a particular mediator complex might be composed of, but it certainly does not accurately depict the conformation of the complex ''in vivo''. During evolution, mediator has become more complex. The yeast ''Saccharomyces cerevisiae'' (a simple eukaryote) is thought to have up to 21 subunits in the core mediator (exclusive of the CDK module), while mammals have up to 26. Individual subunits can be absent or replaced by other subunits under differe ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Kinase
In biochemistry, a kinase () is an enzyme that catalyzes the transfer of phosphate groups from high-energy, phosphate-donating molecules to specific substrates. This process is known as phosphorylation, where the high-energy ATP molecule donates a phosphate group to the substrate molecule. This transesterification produces a phosphorylated substrate and ADP. Conversely, it is referred to as dephosphorylation when the phosphorylated substrate donates a phosphate group and ADP gains a phosphate group (producing a dephosphorylated substrate and the high energy molecule of ATP). These two processes, phosphorylation and dephosphorylation, occur four times during glycolysis. Kinases are part of the larger family of phosphotransferases. Kinases should not be confused with phosphorylases, which catalyze the addition of inorganic phosphate groups to an acceptor, nor with phosphatases, which remove phosphate groups (dephosphorylation). The phosphorylation state of a molecule, whet ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
FG Syndrome
FG syndrome (FGS) is a rare genetic syndrome caused by one or more recessive genes located on the X chromosome and causing physical anomalies and developmental delays. FG syndrome was named after the first letters of the surnames of the first patients noted with the disease. First reported by American geneticists John M. Opitz and Elisabeth G. Kaveggia in 1974, its major clinical features include intellectual disability, hyperactivity, hypotonia (low muscle tone), and a characteristic facial appearance including macrocephaly (an abnormally large head). Presentation FG syndrome's major clinical features include physical disability, usually mild; hyperactive behavior, often with a slow personality; severe constipation, with or without structural anomalies in the anus such as imperforate anus; macrocephaly; severe hypotonia; a characteristic facial appearance due to hypotonia, giving a droopy, "open-mouthed" expression, a thin upper lip, a full or pouting upper lip; and most or com ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Non-coding RNA
A non-coding RNA (ncRNA) is a functional RNA molecule that is not translated into a protein. The DNA sequence from which a functional non-coding RNA is transcribed is often called an RNA gene. Abundant and functionally important types of non-coding RNAs include transfer RNAs (tRNAs) and ribosomal RNAs (rRNAs), as well as small RNAs such as microRNAs, siRNAs, piRNAs, snoRNAs, snRNAs, exRNAs, scaRNAs and the long ncRNAs such as Xist and HOTAIR. The number of non-coding RNAs within the human genome is unknown; however, recent transcriptomic and bioinformatic studies suggest that there are thousands of non-coding transcripts. Many of the newly identified ncRNAs have not been validated for their function. There is no consensus in the literature on how much of non-coding transcription is functional. Some researchers have argued that many ncRNAs are non-functional (sometimes referred to as "junk RNA"), spurious transcriptions. Others, however, disagree, arguing instead that many ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Gene Duplication
Gene duplication (or chromosomal duplication or gene amplification) is a major mechanism through which new genetic material is generated during molecular evolution. It can be defined as any duplication of a region of DNA that contains a gene. Gene duplications can arise as products of several types of errors in DNA replication and repair machinery as well as through fortuitous capture by selfish genetic elements. Common sources of gene duplications include ectopic recombination, retrotransposition event, aneuploidy, polyploidy, and replication slippage. Mechanisms of duplication Ectopic recombination Duplications arise from an event termed unequal crossing-over that occurs during meiosis between misaligned homologous chromosomes. The chance of it happening is a function of the degree of sharing of repetitive elements between two chromosomes. The products of this recombination are a duplication at the site of the exchange and a reciprocal deletion. Ectopic recombination is ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cyclin
Cyclin is a family of proteins that controls the progression of a cell through the cell cycle by activating cyclin-dependent kinase (CDK) enzymes or group of enzymes required for synthesis of cell cycle. Etymology Cyclins were originally discovered by R. Timothy Hunt in 1982 while studying the cell cycle of sea urchins. In an interview for "The Life Scientific" (aired on 13/12/2011) hosted by Jim Al-Khalili, R. Timothy Hunt explained that the name "cyclin" was originally named after his hobby cycling. It was only after the naming did its importance in the cell cycle become apparent. As it was appropriate the name stuck. R. Timothy Hunt: "By the way, the name cyclin, which I coined, was really a joke, it's because I liked cycling so much at the time, but they did come and go in the cell..." Function Cyclins were originally named because their concentration varies in a cyclical fashion during the cell cycle. (Note that the cyclins are now classified according to their conse ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Paralogues
Sequence homology is the biological homology between DNA, RNA, or protein sequences, defined in terms of shared ancestry in the evolutionary history of life. Two segments of DNA can have shared ancestry because of three phenomena: either a speciation event (orthologs), or a duplication event (paralogs), or else a horizontal (or lateral) gene transfer event (xenologs). Homology among DNA, RNA, or proteins is typically inferred from their nucleotide or amino acid sequence similarity. Significant similarity is strong evidence that two sequences are related by evolutionary changes from a common ancestral sequence. Alignments of multiple sequences are used to indicate which regions of each sequence are homologous. Identity, similarity, and conservation The term "percent homology" is often used to mean "sequence similarity”, that is the percentage of identical residues (''percent identity''), or the percentage of residues conserved with similar physicochemical properties ('' ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
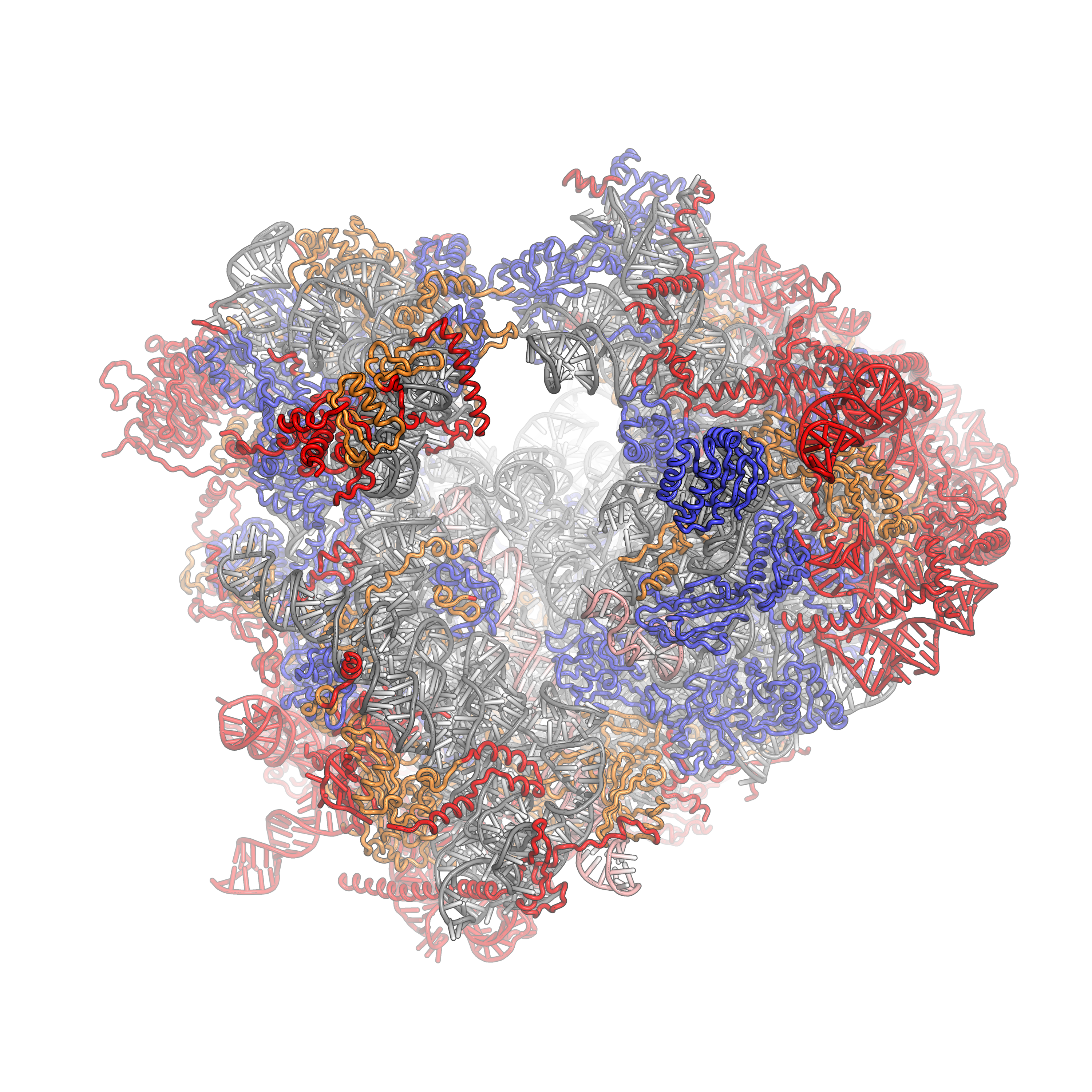
Eukaryotic Ribosome (80S)
Ribosomes are a large and complex molecular machine that catalyzes the synthesis of proteins, referred to as translation. The ribosome selects aminoacylated transfer RNAs (tRNAs) based on the sequence of a protein-encoding messenger RNA (mRNA) and covalently links the amino acids into a polypeptide chain. Ribosomes from all organisms share a highly conserved catalytic center. However, the ribosomes of eukaryotes (animals, plants, fungi, and large number unicellular organisms all with a nucleus) are much larger than prokaryotic (bacterial and archaeal) ribosomes and subject to more complex regulation and biogenesis pathways. Eukaryotic ribosomes are also known as 80S ribosomes, referring to their sedimentation coefficients in Svedberg units, because they sediment faster than the prokaryotic (70S) ribosomes. Eukaryotic ribosomes have two unequal subunits, designated small subunit (40S) and large subunit (60S) according to their sedimentation coefficients. Both subunits contain doze ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Transcription Factor
In molecular biology, a transcription factor (TF) (or sequence-specific DNA-binding factor) is a protein that controls the rate of transcription of genetic information from DNA to messenger RNA, by binding to a specific DNA sequence. The function of TFs is to regulate—turn on and off—genes in order to make sure that they are expressed in the desired cells at the right time and in the right amount throughout the life of the cell and the organism. Groups of TFs function in a coordinated fashion to direct cell division, cell growth, and cell death throughout life; cell migration and organization (body plan) during embryonic development; and intermittently in response to signals from outside the cell, such as a hormone. There are up to 1600 TFs in the human genome. Transcription factors are members of the proteome as well as regulome. TFs work alone or with other proteins in a complex, by promoting (as an activator), or blocking (as a repressor) the recruitment of RNA ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Holoenzyme
Enzymes () are proteins that act as biological catalysts by accelerating chemical reactions. The molecules upon which enzymes may act are called substrates, and the enzyme converts the substrates into different molecules known as product (chemistry), products. Almost all metabolism, metabolic processes in the cell (biology), cell need enzyme catalysis in order to occur at rates fast enough to sustain life. Metabolic pathways depend upon enzymes to catalyze individual steps. The study of enzymes is called ''enzymology'' and the field of pseudoenzyme, pseudoenzyme analysis recognizes that during evolution, some enzymes have lost the ability to carry out biological catalysis, which is often reflected in their amino acid sequences and unusual 'pseudocatalytic' properties. Enzymes are known to catalyze more than 5,000 biochemical reaction types. Other biocatalysts are Ribozyme, catalytic RNA molecules, called ribozymes. Enzymes' Chemical specificity, specificity comes from their u ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |