|
Multicopy Single-stranded DNA
Multicopy single-stranded DNA (msDNA) is a type of extrachromosomal satellite DNA that consists of a single-stranded DNA molecule covalently linked via a 2'-5'phosphodiester bond to an internal guanosine of an RNA molecule. The resultant DNA/RNA chimera possesses two stem-loops joined by a branch similar to the branches found in RNA splicing intermediates. The coding region for msDNA, called a "retron", also encodes a type of reverse transcriptase, which is essential for msDNA synthesis. Discovery Before the discovery of msDNA in myxobacteria, a group of swarming, soil-dwelling bacteria, it was thought that the enzymes known as reverse transcriptases (RT) existed only in eukaryotes and viruses. The discovery led to an increase in research of the area. As a result, msDNA has been found to be widely distributed among bacteria, including various strains of ''Escherichia coli'' and pathogenic bacteria. Further research discovered similarities between HIV-encoded reverse transcriptase ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Bacteriophage
A bacteriophage (), also known informally as a ''phage'' (), is a duplodnaviria virus that infects and replicates within bacteria and archaea. The term was derived from "bacteria" and the Greek φαγεῖν ('), meaning "to devour". Bacteriophages are composed of proteins that encapsulate a DNA or RNA genome, and may have structures that are either simple or elaborate. Their genomes may encode as few as four genes (e.g. MS2) and as many as hundreds of genes. Phages replicate within the bacterium following the injection of their genome into its cytoplasm. Bacteriophages are among the most common and diverse entities in the biosphere. Bacteriophages are ubiquitous viruses, found wherever bacteria exist. It is estimated there are more than 1031 bacteriophages on the planet, more than every other organism on Earth, including bacteria, combined. Viruses are the most abundant biological entity in the water column of the world's oceans, and the second largest component of biom ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Steric Hindrance
Steric effects arise from the spatial arrangement of atoms. When atoms come close together there is a rise in the energy of the molecule. Steric effects are nonbonding interactions that influence the shape ( conformation) and reactivity of ions and molecules. Steric effects complement electronic effects, which dictate the shape and reactivity of molecules. Steric repulsive forces between overlapping electron clouds result in structured groupings of molecules stabilized by the way that opposites attract and like charges repel. Steric hindrance Steric hindrance is a consequence of steric effects. Steric hindrance is the slowing of chemical reactions due to steric bulk. It is usually manifested in ''intermolecular reactions'', whereas discussion of steric effects often focus on ''intramolecular interactions''. Steric hindrance is often exploited to control selectivity, such as slowing unwanted side-reactions. Steric hindrance between adjacent groups can also affect torsional ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DNA Polymerase
A DNA polymerase is a member of a family of enzymes that catalyze the synthesis of DNA molecules from nucleoside triphosphates, the molecular precursors of DNA. These enzymes are essential for DNA replication and usually work in groups to create two identical DNA duplexes from a single original DNA duplex. During this process, DNA polymerase "reads" the existing DNA strands to create two new strands that match the existing ones. These enzymes catalyze the chemical reaction : deoxynucleoside triphosphate + DNAn pyrophosphate + DNAn+1. DNA polymerase adds nucleotides to the three prime (3')-end of a DNA strand, one nucleotide at a time. Every time a cell divides, DNA polymerases are required to duplicate the cell's DNA, so that a copy of the original DNA molecule can be passed to each daughter cell. In this way, genetic information is passed down from generation to generation. Before replication can take place, an enzyme called helicase unwinds the DNA molecule from its tightl ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
CDNA
In genetics, complementary DNA (cDNA) is DNA synthesized from a single-stranded RNA (e.g., messenger RNA (mRNA) or microRNA (miRNA)) template in a reaction catalyzed by the enzyme reverse transcriptase. cDNA is often used to express a specific protein in a cell that does not normally express that protein (i.e., heterologous expression), or to sequence or quantify mRNA molecules using DNA based methods (qPCR, RNA-seq). cDNA that codes for a specific protein can be transferred to a recipient cell for expression, often bacterial or yeast expression systems. cDNA is also generated to analyze transcriptomic profiles in bulk tissue, single cells, or single nuclei in assays such as microarrays, qPCR, and RNA-seq. cDNA is also produced naturally by retroviruses (such as HIV-1, HIV-2, simian immunodeficiency virus, etc.) and then integrated into the host's genome, where it creates a provirus. The term ''cDNA'' is also used, typically in a bioinformatics context, to refer to an mR ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ribozyme
Ribozymes (ribonucleic acid enzymes) are RNA molecules that have the ability to catalyze specific biochemical reactions, including RNA splicing in gene expression, similar to the action of protein enzymes. The 1982 discovery of ribozymes demonstrated that RNA can be both genetic material (like DNA) and a biological catalysis, catalyst (like protein enzymes), and contributed to the RNA world hypothesis, which suggests that RNA may have been important in the evolution of prebiotic self-replicating systems. The most common activities of natural or in vitro-evolved ribozymes are the cleavage or ligation of RNA and DNA and peptide bond formation. For example, the smallest ribozyme known (GUGGC-3') can aminoacylate a GCCU-3' sequence in the presence of PheAMP. Within the ribosome, ribozymes function as part of the large subunit ribosomal RNA to link amino acids during Translation (biology), protein synthesis. They also participate in a variety of RNA processing reactions, including RNA ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Spliceosome
A spliceosome is a large ribonucleoprotein (RNP) complex found primarily within the nucleus of eukaryotic cells. The spliceosome is assembled from small nuclear RNAs (snRNA) and numerous proteins. Small nuclear RNA (snRNA) molecules bind to specific proteins to form a small nuclear ribonucleoprotein complex (snRNP, pronounced “snurps”), which in turn combines with other snRNPs to form a large ribonucleoprotein complex called a spliceosome. The spliceosome removes introns from a transcribed pre-mRNA, a type of primary transcript. This process is generally referred to as splicing. An analogy is a film editor, who selectively cuts out irrelevant or incorrect material (equivalent to the introns) from the initial film and sends the cleaned-up version to the director for the final cut. However, sometimes the RNA within the intron acts as a ribozyme, splicing itself without the use of a spliceosome or protein enzymes. History In 1977, work by the Sharp and Roberts labs reveale ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Msdna Synthesis
Multicopy single-stranded DNA (msDNA) is a type of extrachromosomal satellite DNA that consists of a single-stranded DNA molecule covalently linked via a 2'-5'phosphodiester bond to an internal guanosine of an RNA molecule. The resultant DNA/RNA chimera possesses two stem-loops joined by a branch similar to the branches found in RNA splicing intermediates. The coding region for msDNA, called a "retron", also encodes a type of reverse transcriptase, which is essential for msDNA synthesis. Discovery Before the discovery of msDNA in myxobacteria, a group of swarming, soil-dwelling bacteria, it was thought that the enzymes known as reverse transcriptases (RT) existed only in eukaryotes and viruses. The discovery led to an increase in research of the area. As a result, msDNA has been found to be widely distributed among bacteria, including various strains of ''Escherichia coli'' and pathogenic bacteria. Further research discovered similarities between HIV-encoded reverse transcriptas ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Allorecognition
Allorecognition is the ability of an individual organism to distinguish its own tissues from those of another. It manifests itself in the recognition of antigens expressed on the surface of cells of non-self origin. Allorecognition has been described in nearly all multicellular phyla. This article focuses on allorecognition from the standpoint of its significance in the evolution of multicellular organisms. For other articles which focus on its importance in medicine, molecular biology, and so forth, the following topics are recommended as well as those in the Categories links at the bottom of this page. * Immune system, Immunology * Transplant rejection * Tissue typing * Major histocompatibility complex (MHC) The ability to discriminate between self and non-self is a fundamental requirement for life. At the most basic level, even single-celled organisms need to be able to distinguish between food and non-food, to respond appropriately to invading pathogens, and to avoid cannibal ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
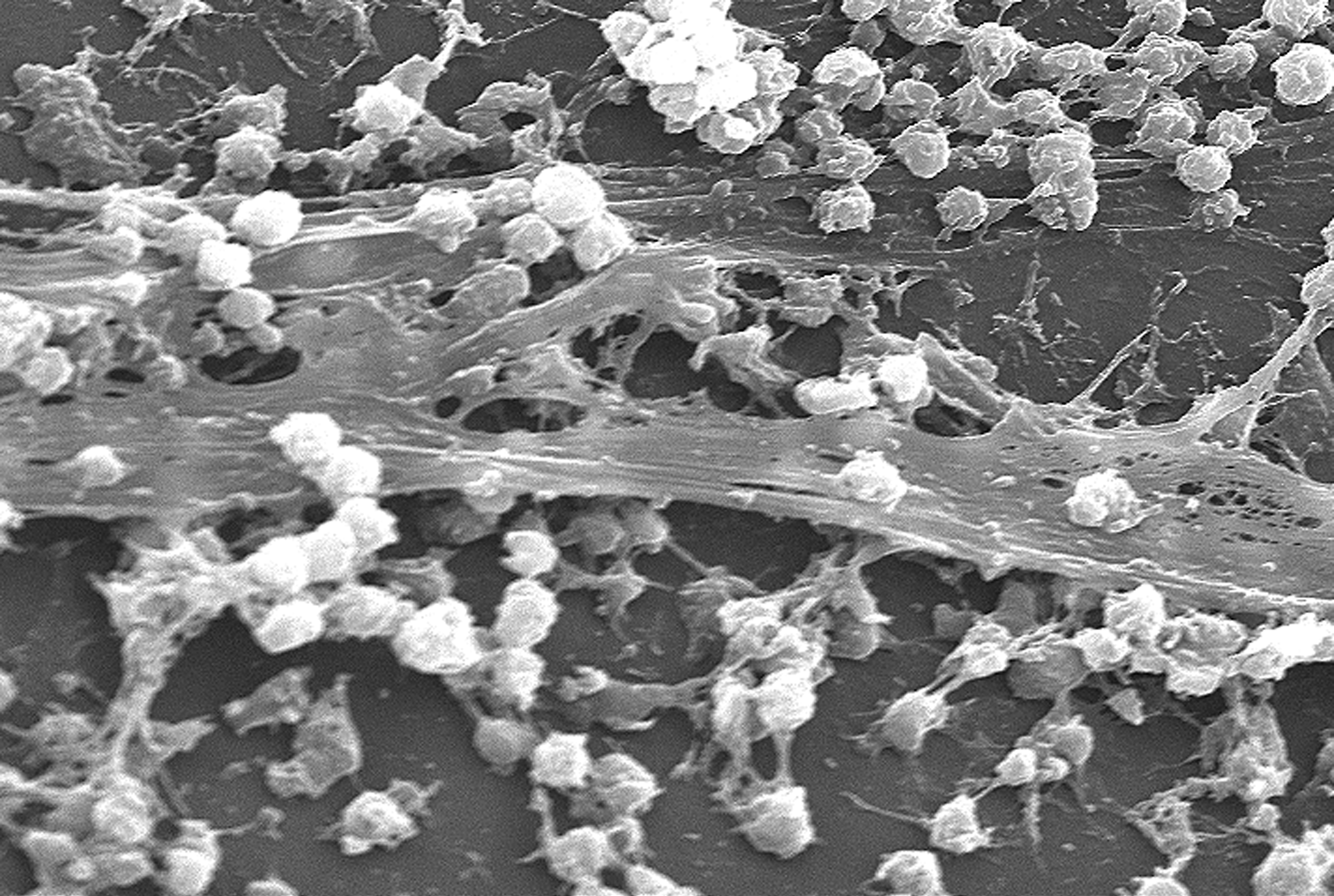
Biofilm
A biofilm comprises any syntrophic consortium of microorganisms in which cells stick to each other and often also to a surface. These adherent cells become embedded within a slimy extracellular matrix that is composed of extracellular polymeric substances (EPSs). The cells within the biofilm produce the EPS components, which are typically a polymeric conglomeration of extracellular polysaccharides, proteins, lipids and DNA. Because they have three-dimensional structure and represent a community lifestyle for microorganisms, they have been metaphorically described as "cities for microbes". Biofilms may form on living or non-living surfaces and can be prevalent in natural, industrial, and hospital settings. They may constitute a microbiome or be a portion of it. The microbial cells growing in a biofilm are physiologically distinct from planktonic cells of the same organism, which, by contrast, are single cells that may float or swim in a liquid medium. Biofilms can form ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Sequence Homology
Sequence homology is the biological homology between DNA, RNA, or protein sequences, defined in terms of shared ancestry in the evolutionary history of life. Two segments of DNA can have shared ancestry because of three phenomena: either a speciation event (orthologs), or a duplication event (paralogs), or else a horizontal (or lateral) gene transfer event (xenologs). Homology among DNA, RNA, or proteins is typically inferred from their nucleotide or amino acid sequence similarity. Significant similarity is strong evidence that two sequences are related by evolutionary changes from a common ancestral sequence. Alignments of multiple sequences are used to indicate which regions of each sequence are homologous. Identity, similarity, and conservation The term "percent homology" is often used to mean "sequence similarity”, that is the percentage of identical residues (''percent identity''), or the percentage of residues conserved with similar physicochemical properties (' ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Stigmatella Aurantiaca
''Stigmatella aurantiaca'' is a member of myxobacteria, a group of gram-negative bacteria with a complex developmental life cycle. Classification The bacterial nature of this organism was recognized by Thaxter in 1892, who grouped it among the ''Chrondromyces''. It had been described several times before, but had been misclassified as a member of the ''fungi imperfecti''. More recent investigations have shown that, contrary to Thaxter's classification, this organism is not closely related to ''Chrondromyces'', and ''Stigmatella'' is currently recognized as a separate genus. Of the three major subgroups of the myxobacteria, Myxococcus, Nannocystis, and Chrondromyces, ''Stigmatella'' is most closely aligned with Myxococcus. Life cycle ''S. aurantiaca'', like other myxobacterial species, has a complex life cycle including social gliding (swarming), fruiting body formation, and predatory feeding behaviors. The bacteria do not swim, but glide on surfaces leaving slime trails ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |