|
Locus Heterogeneity
Locus heterogeneity occurs when mutations at multiple genomic loci are capable of producing the same phenotype (ie. a single trait, pattern of traits, or disorder), and each individual mutation is sufficient to cause the specific phenotype independently. Locus heterogeneity should not be confused with allelic heterogeneity, in which a single phenotype can be produced by multiple mutations, all of which are at the same locus on a chromosome. Likewise, it should not be confused with phenotypic heterogeneity, in which different phenotypes arise among organisms with identical genotypes and environmental conditions. Locus heterogeneity and allelic heterogeneity are the two components of genetic heterogeneity. Locus heterogeneity may have major implications for a number of human diseases. For instance, it has been associated with retinitis pigmentosa, hypertrophic cardiomyopathy, osteogenesis imperfecta, and familial hypercholesterolemia. Heterogenous loci involved in formation of the ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Phenotype
In genetics, the phenotype () is the set of observable characteristics or traits of an organism. The term covers the organism's morphology or physical form and structure, its developmental processes, its biochemical and physiological properties, its behavior, and the products of behavior. An organism's phenotype results from two basic factors: the expression of an organism's genetic code, or its genotype, and the influence of environmental factors. Both factors may interact, further affecting phenotype. When two or more clearly different phenotypes exist in the same population of a species, the species is called polymorphic. A well-documented example of polymorphism is Labrador Retriever coloring; while the coat color depends on many genes, it is clearly seen in the environment as yellow, black, and brown. Richard Dawkins in 1978 and then again in his 1982 book '' The Extended Phenotype'' suggested that one can regard bird nests and other built structures such as ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
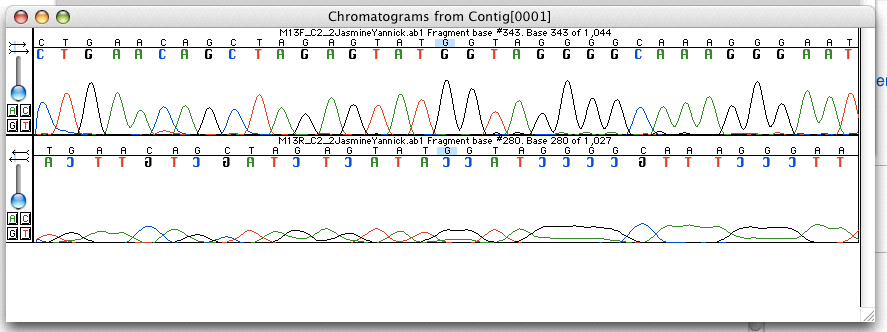
Whole Genome Sequencing
Whole genome sequencing (WGS), also known as full genome sequencing, complete genome sequencing, or entire genome sequencing, is the process of determining the entirety, or nearly the entirety, of the DNA sequence of an organism's genome at a single time. This entails sequencing all of an organism's chromosomal DNA as well as DNA contained in the mitochondria and, for plants, in the chloroplast. Whole genome sequencing has largely been used as a research tool, but was being introduced to clinics in 2014. In the future of personalized medicine, whole genome sequence data may be an important tool to guide therapeutic intervention. The tool of gene sequencing at SNP level is also used to pinpoint functional variants from association studies and improve the knowledge available to researchers interested in evolutionary biology, and hence may lay the foundation for predicting disease susceptibility and drug response. Whole genome sequencing should not be confused with DNA profiling ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Allelic Heterogeneity
Allelic heterogeneity is the phenomenon in which different mutations at the same locus lead to the same or very similar phenotypes. These allelic variations can arise as a result of natural selection processes, as a result of exogenous mutagens, genetic drift, or genetic migration. Many of these mutations take the form of single nucleotide polymorphisms in which a single nucleotide base is altered compared to a consensus sequence. They can also exist as copy number variants (CNV) in which the copies of a gene or DNA sequence is different from the population. Mutated alleles expressing allelic heterogeneity can be classified as adaptive or disadaptive. These mutations can occur in the germ line cells, somatic cells, or in the mitochondrial. Mutations in germ line cells can be inherited as well as mitochondrial allelic mutations. The mitochondrial allelic mutations are inherited maternally. Typically in the human genome a small amount of allele variants account for ~75% of the ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Retinitis Pigmentosa GTPase Regulator
X-linked retinitis pigmentosa GTPase regulator is a GTPase-binding protein that in humans is encoded by the ''RPGR'' gene. The gene is located on the X-chromosome and is commonly associated with X-linked retinitis pigmentosa (XLRP). In photoreceptor cells, RPGR is localized in the connecting cilium which connects the protein-synthesizing inner segment to the photosensitive outer segment and is involved in the modulation of cargo trafficked between the two segments. Function This gene encodes a protein with a series of six RCC1-like domains (RLDs), characteristic of the highly conserved guanine nucleotide exchange factors. Mutations in this gene have been associated with X-linked retinitis pigmentosa (XLRP). Multiple alternatively spliced transcript variants that encode different isoforms of this gene have been reported, but the full-length natures of only some have been determined. The two major isoforms are RPGRconst, the default isoform, composed of exons 1-19, and RPGRORF15 ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Rhodopsin
Rhodopsin, also known as visual purple, is a protein encoded by the RHO gene and a G-protein-coupled receptor (GPCR). It is the opsin of the rod cells in the retina and a light-sensitive receptor protein that triggers visual phototransduction in rods. Rhodopsin mediates dim light vision and thus is extremely sensitive to light. When rhodopsin is exposed to light, it immediately photobleaches. In humans, it is regenerated fully in about 30 minutes, after which the rods are more sensitive. Defects in the rhodopsin gene cause eye diseases such as retinitis pigmentosa and congenital stationary night blindness. Names Rhodopsin was discovered by Franz Christian Boll in 1876. The name rhodospsin derives from Ancient Greek () for "rose", due to its pinkish color, and () for "sight". It was coined in 1878 by the German physiologist Wilhelm Friedrich Kühne (1837-1900). When George Wald discovered that rhodopsin is a holoprotein, consisting of retinal and an apoprotein, ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Sex Linkage
Sex linked describes the sex-specific patterns of inheritance and presentation when a gene mutation (allele) is present on a sex chromosome (allosome) rather than a non-sex chromosome (autosome). In humans, these are termed X-linked recessive, X-linked dominant and Y-linked. The inheritance and presentation of all three differ depending on the sex of both the parent and the child. This makes them characteristically different from autosomal dominance and recessiveness. There are many more X-linked conditions than Y-linked conditions, since humans have several times as many genes on the X chromosome than the Y chromosome. Only females are able to be carriers for X-linked conditions; males will always be affected by any X-linked condition, since they have no second X chromosome with a healthy copy of the gene. As such, X-linked recessive conditions affect males much more commonly than females. In X-linked recessive inheritance, a son born to a carrier mother and an unaffected f ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Genetic Disorder
A genetic disorder is a health problem caused by one or more abnormalities in the genome. It can be caused by a mutation in a single gene (monogenic) or multiple genes (polygenic) or by a chromosomal abnormality. Although polygenic disorders are the most common, the term is mostly used when discussing disorders with a single genetic cause, either in a gene or chromosome. The mutation responsible can occur spontaneously before embryonic development (a ''de novo'' mutation), or it can be inherited from two parents who are carriers of a faulty gene (autosomal recessive inheritance) or from a parent with the disorder (autosomal dominant inheritance). When the genetic disorder is inherited from one or both parents, it is also classified as a hereditary disease. Some disorders are caused by a mutation on the X chromosome and have X-linked inheritance. Very few disorders are inherited on the Y chromosome or mitochondrial DNA (due to their size). There are well over 6,000 known genet ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Retinitis Pigmentosa
Retinitis pigmentosa (RP) is a genetic disorder of the eyes that causes loss of vision. Symptoms include trouble seeing at night and decreasing peripheral vision (side and upper or lower visual field). As peripheral vision worsens, people may experience " tunnel vision". Complete blindness is uncommon. Onset of symptoms is generally gradual and often begins in childhood. Retinitis pigmentosa is generally inherited from one or both parents or rarely it can be caused by a miscoding during DNA division. It is caused by genetic miscoding of proteins in one of more than 300 genes involved. The underlying mechanism involves the progressive loss of rod photoreceptor cells that line the retina of the eyeball. The rod cells secrete a neuroprotective substance (Rod-derived cone viability factor, RdCVF) that protects the cone cells from apoptosis (cell death). However, when the rod cells die, this substance is no longer provided. This is generally followed by the loss of cone photorece ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Genetic Linkage
Genetic linkage is the tendency of DNA sequences that are close together on a chromosome to be inherited together during the meiosis phase of sexual reproduction. Two genetic markers that are physically near to each other are unlikely to be separated onto different chromatids during chromosomal crossover, and are therefore said to be more ''linked'' than markers that are far apart. In other words, the nearer two genes are on a chromosome, the lower the chance of recombination between them, and the more likely they are to be inherited together. Markers on different chromosomes are perfectly ''unlinked'', although the penetrance of potentially deleterious alleles may be influenced by the presence of other alleles, and these other alleles may be located on other chromosomes than that on which a particular potentially deleterious allele is located. Genetic linkage is the most prominent exception to Gregor Mendel's Law of Independent Assortment. The first experiment to demonstra ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Allelic Heterogeneity
Allelic heterogeneity is the phenomenon in which different mutations at the same locus lead to the same or very similar phenotypes. These allelic variations can arise as a result of natural selection processes, as a result of exogenous mutagens, genetic drift, or genetic migration. Many of these mutations take the form of single nucleotide polymorphisms in which a single nucleotide base is altered compared to a consensus sequence. They can also exist as copy number variants (CNV) in which the copies of a gene or DNA sequence is different from the population. Mutated alleles expressing allelic heterogeneity can be classified as adaptive or disadaptive. These mutations can occur in the germ line cells, somatic cells, or in the mitochondrial. Mutations in germ line cells can be inherited as well as mitochondrial allelic mutations. The mitochondrial allelic mutations are inherited maternally. Typically in the human genome a small amount of allele variants account for ~75% of the ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Familial Hypercholesterolemia
Familial hypercholesterolemia (FH) is a genetic disorder characterized by high cholesterol levels, specifically very high levels of low-density lipoprotein (LDL cholesterol), in the blood and early cardiovascular disease. The most common mutations diminish the number of functional LDL receptors in the liver. Since the underlying body biochemistry is slightly different in individuals with FH, their high cholesterol levels are less responsive to the kinds of cholesterol control methods which are usually more effective in people without FH (such as dietary modification and statin tablets). Nevertheless, treatment (including higher statin doses) is usually effective. FH is classified as a type 2 familial dyslipidemia. There are five types of familial dyslipidemia (not including subtypes), and each are classified from both the altered lipid profile and by the genetic abnormality. For example, high LDL (often due to LDL receptor defect) is type 2. Others include defects in chylomicr ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Osteogenesis Imperfecta
Osteogenesis imperfecta (; OI), colloquially known as brittle bone disease, is a group of genetic disorders that all result in bones that break easily. The range of symptoms—on the skeleton as well as on the body's other organs—may be mild to severe. Symptoms found in various types of OI include whites of the eye (sclerae) that are blue instead, short stature, loose joints, hearing loss, breathing problems and problems with the teeth (dentinogenesis imperfecta). Potentially life-threatening complications, all of which become more common in more severe OI, include: tearing ( dissection) of the major arteries, such as the aorta; pulmonary valve insufficiency secondary to distortion of the ribcage; and basilar invagination. The underlying mechanism is usually a problem with connective tissue due to a lack of, or poorly formed, type I collagen. In more than 90% of cases, OI occurs due to mutations in the ''COL1A1'' or '' COL1A2'' genes. These mutations may be i ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |