|
Lambda Phage
''Enterobacteria phage λ'' (lambda phage, coliphage λ, officially ''Escherichia virus Lambda'') is a bacterial virus, or bacteriophage, that infects the bacterial species ''Escherichia coli'' (''E. coli''). It was discovered by Esther Lederberg in 1950. The wild type of this virus has a temperate life cycle that allows it to either reside within the genome of its host through lysogeny or enter into a lytic phase, during which it kills and lyses the cell to produce offspring. Lambda strains, mutated at specific sites, are unable to lysogenize cells; instead, they grow and enter the lytic cycle after superinfecting an already lysogenized cell. The phage particle consists of a head (also known as a capsid), a tail, and tail fibers (see image of virus below). The head contains the phage's double-strand linear DNA genome. During infection, the phage particle recognizes and binds to its host, ''E. coli'', causing DNA in the head of the phage to be ejected through the tail into the ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Lambda 7-24-2021 Ps
Lambda (}, ''lám(b)da'') is the 11th letter of the Greek alphabet, representing the Dental, alveolar and postalveolar lateral approximants, voiced alveolar lateral approximant . In the system of Greek numerals, lambda has a value of 30. Lambda is derived from the Phoenician alphabet, Phoenician Lamed . Lambda gave rise to the Latin script, Latin L and the Cyrillic script, Cyrillic El (Cyrillic), El (Л). The ancient Alexandrine grammarians, grammarians and dramatists give evidence to the pronunciation as () in Classical Greek times. In Modern Greek, the name of the letter, Λάμδα, is pronounced . In Epichoric alphabets, early Greek alphabets, the shape and orientation of lambda varied. Most variants consisted of two straight strokes, one longer than the other, connected at their ends. The angle might be in the upper-left, lower-left ("Western" alphabets) or top ("Eastern" alphabets). Other variants had a vertical line with a horizontal or sloped stroke running to the right. ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Porin (protein)
Porins are beta barrel proteins that cross a cellular membrane and act as a pore, through which molecules can diffuse. Unlike other membrane transport proteins, porins are large enough to allow passive diffusion, i.e., they act as channels that are specific to different types of molecules. They are present in the outer membrane of gram-negative bacteria and some gram-positive mycobacteria (mycolic acid-containing actinomycetes), the outer membrane of mitochondria, and the outer chloroplast membrane (outer plastid membrane). Structure Porins are composed of beta sheets (β sheets) made up of beta strands (β strands) which are linked together by beta turns on the cytoplasmic side and long loops of amino acids on the other. The β strands lie in an antiparallel fashion and form a cylindrical tube, called a beta barrel (β barrel). The amino acid composition of the porin β strands are unique in that polar and nonpolar residues alternate along them. This means that the nonpolar ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Multiplicity Of Infection
In microbiology, the multiplicity of infection or MOI is the ratio of agents (e.g. phage or more generally virus, bacteria) to infection targets (e.g. cell). For example, when referring to a group of cells inoculated with virus particles, the MOI is the ratio of the number of virus particles to the number of target cells present in a defined space. Interpretation The actual number of viruses or bacteria that will enter any given cell is a stochastic process: some cells may absorb more than one infectious agent, while others may not absorb any. Before determining the multiplicity of infection, it's absolutely necessary to have a well-isolated agent, as crude agents may not produce reliable and reproducible results. The probability that a cell will absorb n virus particles or bacteria when inoculated with an MOI of m can be calculated for a given population using a Poisson distribution. This application of Poisson's distribution was applied and described by Ellis and Delbrück. : P(n ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
CII Protein
cII or transcriptional activator II is a DNA-binding protein and important transcription factor in the life cycle of lambda phage. It is encoded in the lambda phage genome by the 291 base pair cII gene. cII plays a key role in determining whether the bacteriophage will incorporate its genome into its host and lie dormant (lysogeny), or replicate and kill the host ( lysis). Introduction cII is the central “switchman” in the lambda phage bistable genetic switch, allowing environmental and cellular conditions to factor into the decision to lysogenize or to lyse its host. cII acts as a transcriptional activator of three promoters on the phage genome: pI, pRE, and pAQ. cII is an unstable protein with a half-life as short as 1.5 mins at 37˚C, enabling rapid fluctuations in its concentration. First isolated in 1982, cII's function in lambda's regulatory network has been extensively studied. Structure and properties cII binds DNA as a tetramer, composed of identical 11 kDa subuni ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Bacteriostatic
A bacteriostatic agent or bacteriostat, abbreviated Bstatic, is a biological or chemical agent that stops bacteria from reproducing, while not necessarily killing them otherwise. Depending on their application, bacteriostatic antibiotics, disinfectants, antiseptics and preservatives can be distinguished. When bacteriostatic antimicrobials are used, the duration of therapy must be sufficient to allow host defense mechanisms to eradicate the bacteria. Upon removal of the bacteriostat, the bacteria usually start to grow rapidly. This is in contrast to bactericides, which kill bacteria. Bacteriostats are often used in plastics to prevent growth of bacteria on surfaces. Bacteriostats commonly used in laboratory work include sodium azide (which is acutely toxic) and thiomersal. __TOC__ Bacteriostatic antibiotics Bacteriostatic antibiotics limit the growth of bacteria by interfering with bacterial protein production, DNA replication, or other aspects of bacterial cellular metabolism. The ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
RNA Polymerase
In molecular biology, RNA polymerase (abbreviated RNAP or RNApol), or more specifically DNA-directed/dependent RNA polymerase (DdRP), is an enzyme that synthesizes RNA from a DNA template. Using the enzyme helicase, RNAP locally opens the double-stranded DNA so that one strand of the exposed nucleotides can be used as a template for the synthesis of RNA, a process called transcription. A transcription factor and its associated transcription mediator complex must be attached to a DNA binding site called a promoter region before RNAP can initiate the DNA unwinding at that position. RNAP not only initiates RNA transcription, it also guides the nucleotides into position, facilitates attachment and elongation, has intrinsic proofreading and replacement capabilities, and termination recognition capability. In eukaryotes, RNAP can build chains as long as 2.4 million nucleotides. RNAP produces RNA that, functionally, is either for protein coding, i.e. messenger RNA (mRNA); or n ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Antiterminator
Antitermination is the prokaryotic cell's aid to fix premature termination of RNA synthesis during the transcription of RNA. It occurs when the RNA polymerase ignores the termination signal and continues elongating its transcript until a second signal is reached. Antitermination provides a mechanism whereby one or more genes at the end of an operon can be switched either on or off, depending on the polymerase either recognizing or not recognizing the termination signal. Antitermination is used by some phages to regulate progression from one stage of gene expression to the next. The lambda gene N, codes for an antitermination protein (pN) that is necessary to allow RNA polymerase to read through the terminators located at the ends of the immediate early genes. Another antitermination protein, pQ, is required later in phage infection. pN and pQ act on RNA polymerase as it passes specific sites. These sites are located at different relative positions in their respective transcription un ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
N Protien
N, or n, is the fourteenth letter in the Latin alphabet, used in the modern English alphabet, the alphabets of other western European languages and others worldwide. Its name in English is ''en'' (pronounced ), plural ''ens''. History One of the most common hieroglyphs, snake, was used in Egyptian writing to stand for a sound like the English , because the Egyptian word for "snake" was ''djet''. It is speculated by many that Semitic people working in Egypt adapted hieroglyphics to create the first alphabet, and that they used the same snake symbol to represent N, because their word for "snake" may have begun with that sound. However, the name for the letter in the Phoenician, Hebrew, Aramaic and Arabic alphabets is ''nun'', which means "fish" in some of these languages. The sound value of the letter was —as in Greek, Etruscan, Latin and modern languages. Use in writing systems represents a dental or alveolar nasal in virtually all languages that use the Latin alp ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Promoter (biology)
In genetics, a promoter is a sequence of DNA to which proteins bind to initiate transcription of a single RNA transcript from the DNA downstream of the promoter. The RNA transcript may encode a protein (mRNA), or can have a function in and of itself, such as tRNA or rRNA. Promoters are located near the transcription start sites of genes, upstream on the DNA (towards the 5' region of the sense strand). Promoters can be about 100–1000 base pairs long, the sequence of which is highly dependent on the gene and product of transcription, type or class of RNA polymerase recruited to the site, and species of organism. Promoters control gene expression in bacteria and eukaryotes. RNA polymerase must attach to DNA near a gene for transcription to occur. Promoter DNA sequences provide an enzyme binding site. The -10 sequence is TATAAT. -35 sequences are conserved on average, but not in most promoters. Artificial promoters with conserved -10 and -35 elements transcribe more slowly. All D ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Supercoil
DNA supercoiling refers to the amount of twist in a particular DNA strand, which determines the amount of strain on it. A given strand may be "positively supercoiled" or "negatively supercoiled" (more or less tightly wound). The amount of a strand’s supercoiling affects a number of biological processes, such as compacting DNA and regulating access to the genetic code (which strongly affects DNA metabolism and possibly gene expression). Certain enzymes, such as topoisomerases, change the amount of DNA supercoiling to facilitate functions such as DNA replication and transcription. The amount of supercoiling in a given strand is described by a mathematical formula that compares it to a reference state known as "relaxed B-form" DNA. Overview In a "relaxed" double-helical segment of B-DNA, the two strands twist around the helical axis once every 10.4–10.5 base pairs of sequence. Adding or subtracting twists, as some enzymes do, imposes strain. If a DNA segment under twist ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
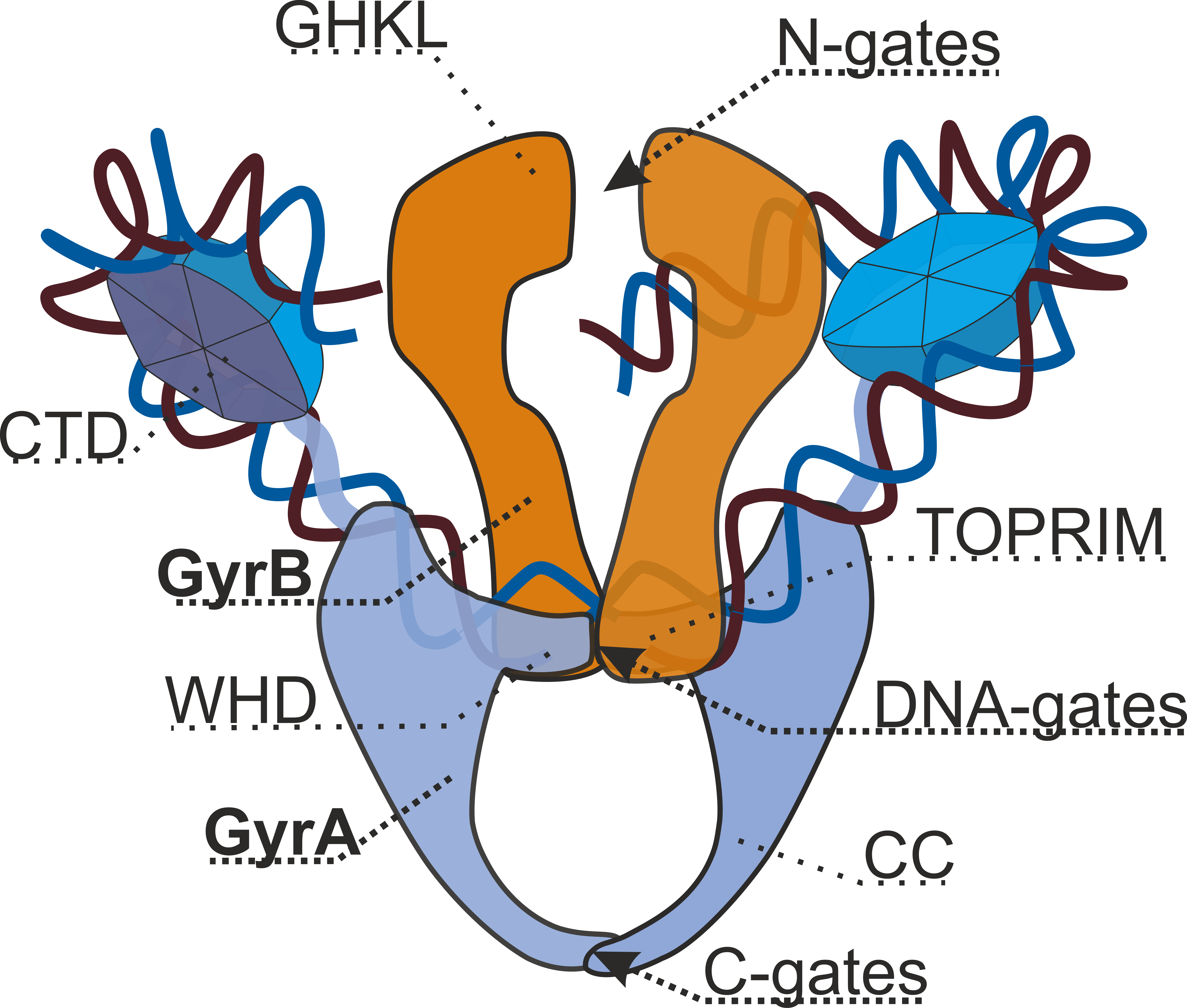
DNA Gyrase
DNA gyrase, or simply gyrase, is an enzyme Enzymes () are proteins that act as biological catalysts by accelerating chemical reactions. The molecules upon which enzymes may act are called substrates, and the enzyme converts the substrates into different molecules known as products ... within the class of topoisomerase and is a subclass of Type II topoisomerases that reduces topological strain in an ATP dependent manner while double-stranded DNA is being unwound by elongating RNA polymerase, RNA-polymerase or by helicase in front of the progressing DNA replication#Replication fork, replication fork. The enzyme causes negative DNA supercoil, supercoiling of the DNA or relaxes positive supercoils. It does so by looping the template so as to form a crossing, then cutting one of the double helices and passing the other through it before releasing the break, changing the linking number by two in each enzymatic step. This process occurs in bacteria, whose single circular DNA is ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |