|
Supercoil
DNA supercoiling refers to the amount of twist in a particular DNA strand, which determines the amount of strain on it. A given strand may be "positively supercoiled" or "negatively supercoiled" (more or less tightly wound). The amount of a strand’s supercoiling affects a number of biological processes, such as compacting DNA and regulating access to the genetic code (which strongly affects DNA metabolism and possibly gene expression). Certain enzymes, such as topoisomerases, change the amount of DNA supercoiling to facilitate functions such as DNA replication and transcription. The amount of supercoiling in a given strand is described by a mathematical formula that compares it to a reference state known as "relaxed B-form" DNA. Overview In a "relaxed" double-helical segment of B-DNA, the two strands twist around the helical axis once every 10.4–10.5 base pairs of sequence. Adding or subtracting twists, as some enzymes do, imposes strain. If a DNA segment under twis ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Topoisomerases
DNA topoisomerases (or topoisomerases) are enzymes that catalyze changes in the topological state of DNA, interconverting relaxed and supercoiled forms, linked (catenated) and unlinked species, and knotted and unknotted DNA. Topological issues in DNA arise due to the intertwined nature of its double-helical structure, which, for example, can lead to overwinding of the DNA duplex during DNA replication and transcription. If left unchanged, this torsion would eventually stop the DNA or RNA polymerases involved in these processes from continuing along the DNA helix. A second topological challenge results from the linking or tangling of DNA during replication. Left unresolved, links between replicated DNA will impede cell division. The DNA topoisomerases prevent and correct these types of topological problems. They do this by binding to DNA and cutting the sugar-phosphate backbone of either one (type I topoisomerases) or both (type II topoisomerases) of the DNA strands. This transien ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
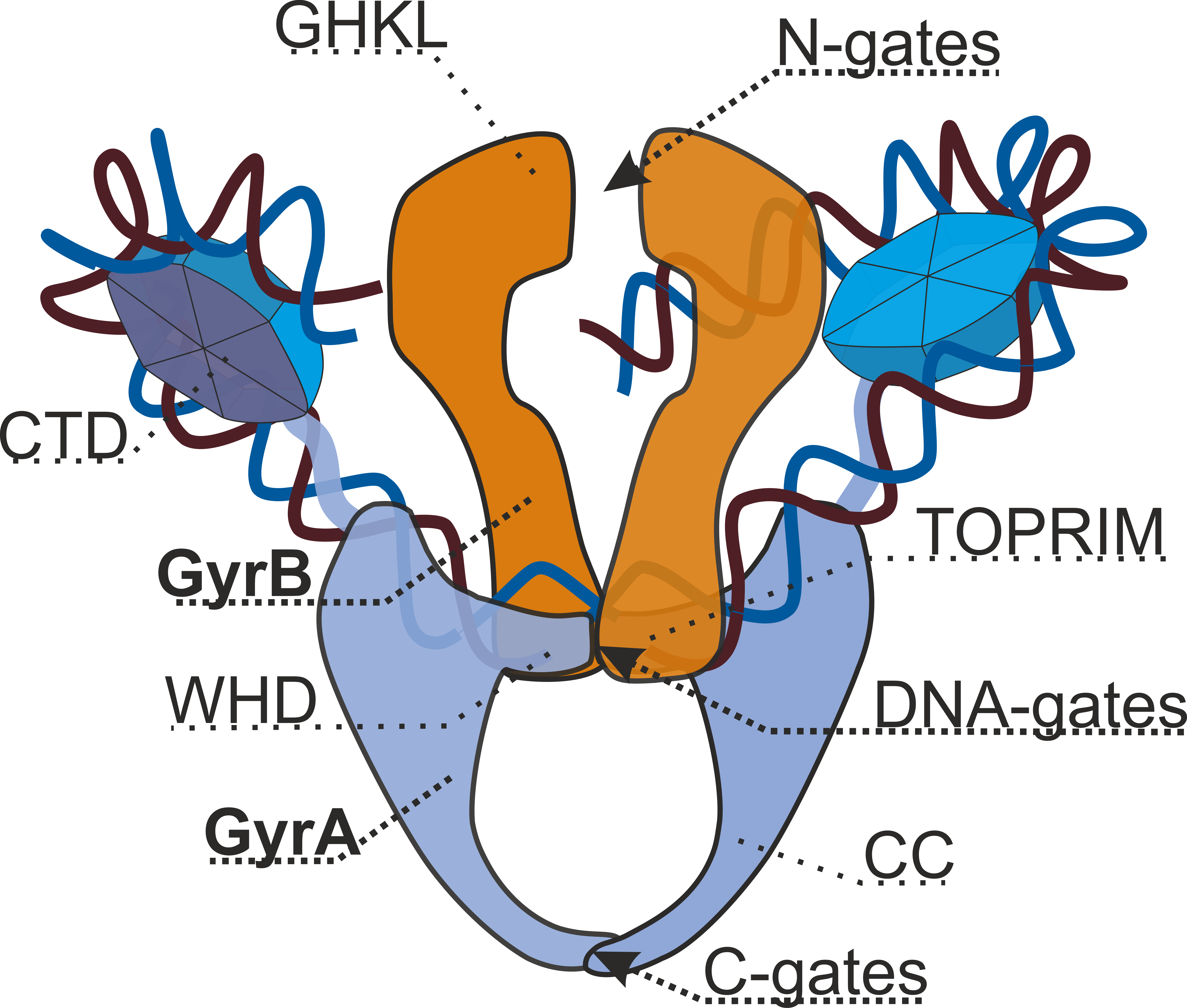
DNA Gyrase
DNA gyrase, or simply gyrase, is an enzyme within the class of topoisomerase and is a subclass of Type II topoisomerases that reduces topological strain in an ATP dependent manner while double-stranded DNA is being unwound by elongating RNA-polymerase or by helicase in front of the progressing replication fork. The enzyme causes negative supercoiling of the DNA or relaxes positive supercoils. It does so by looping the template so as to form a crossing, then cutting one of the double helices and passing the other through it before releasing the break, changing the linking number by two in each enzymatic step. This process occurs in bacteria, whose single circular DNA is cut by DNA gyrase and the two ends are then twisted around each other to form supercoils. Gyrase is also found in eukaryotic plastids: it has been found in the apicoplast of the malarial parasite '' Plasmodium falciparum'' and in chloroplasts of several plants. Bacterial DNA gyrase is the target of many antibi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Condensin
Condensins are large protein complexes that play a central role in chromosome assembly and segregation during mitosis and meiosis (Figure 1). Their subunits were originally identified as major components of mitotic chromosomes assembled in ''Xenopus'' egg extracts. Subunit composition Eukaryotic types Many eukaryotic cells possess two different types of condensin complexes, known as condensin I and condensin II, each of which is composed of five subunits (Figure 2). Condensins I and II share the same pair of core subunits, SMC2 and SMC4, both belonging to a large family of chromosomal ATPases, known as SMC proteins (SMC stands for Structural Maintenance of Chromosomes). Each of the complexes contains a distinct set of non-SMC regulatory subunits (a kleisin subunit and a pair of HEAT repeat subunits). Both complexes are large, having a total molecular mass of 650-700 kDa. The core subunits condensins (SMC2 and SMC4) are conserved among all eukaryotic species that have bee ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DNA Topology
Nucleic acid structure refers to the structure of nucleic acids such as DNA and RNA. Chemically speaking, DNA and RNA are very similar. Nucleic acid structure is often divided into four different levels: primary, secondary, tertiary, and quaternary. Primary structure Primary structure consists of a linear sequence of nucleotides that are linked together by phosphodiester bond. It is this linear sequence of nucleotides that make up the primary structure of DNA or RNA. Nucleotides consist of 3 components: # Nitrogenous base ## Adenine ## Guanine ## Cytosine ## Thymine (present in DNA only) ## Uracil (present in RNA only) # 5-carbon sugar which is called deoxyribose (found in DNA) and ribose (found in RNA). # One or more phosphate groups. The nitrogen bases adenine and guanine are purine in structure and form a glycosidic bond between their 9 nitrogen and the 1' -OH group of the deoxyribose. Cytosine, thymine, and uracil are pyrimidines, hence the glycosidic bonds form betwe ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Topologically Associating Domain
A topologically associating domain (TAD) is a self-interacting genomic region, meaning that DNA sequences within a TAD physically interact with each other more frequently than with sequences outside the TAD. The median size of a TAD in mouse cells is 880 kb, and they have similar sizes in non-mammalian species. Boundaries at both side of these domains are conserved between different mammalian cell types and even across species and are highly enriched with CCCTC-binding factor (CTCF) and cohesin. In addition, some types of genes (such as transfer RNA genes and housekeeping genes) appear near TAD boundaries more often than would be expected by chance. The functions of TADs are not fully understood and are still a matter of debate. Most of the studies indicate TADs regulate gene expression by limiting the enhancer- promoter interaction to each TAD, however, a recent study uncouples TAD organization and gene expression. Disruption of TAD boundaries are found to be associated with w ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Writhe
In knot theory, there are several competing notions of the quantity writhe, or \operatorname. In one sense, it is purely a property of an oriented link diagram and assumes integer values. In another sense, it is a quantity that describes the amount of "coiling" of a mathematical knot (or any closed simple curve) in three-dimensional space and assumes real numbers as values. In both cases, writhe is a geometric quantity, meaning that while deforming a curve (or diagram) in such a way that does not change its topology, one may still change its writhe. Writhe of link diagrams In knot theory, the writhe is a property of an oriented link diagram. The writhe is the total number of positive crossings minus the total number of negative crossings. A direction is assigned to the link at a point in each component and this direction is followed all the way around each component. For each crossing one comes across while traveling in this direction, if the strand underneath goes from right ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Plasmid
A plasmid is a small, extrachromosomal DNA molecule within a cell that is physically separated from chromosomal DNA and can replicate independently. They are most commonly found as small circular, double-stranded DNA molecules in bacteria; however, plasmids are sometimes present in archaea and eukaryotic organisms. In nature, plasmids often carry genes that benefit the survival of the organism and confer selective advantage such as antibiotic resistance. While chromosomes are large and contain all the essential genetic information for living under normal conditions, plasmids are usually very small and contain only additional genes that may be useful in certain situations or conditions. Artificial plasmids are widely used as vectors in molecular cloning, serving to drive the replication of recombinant DNA sequences within host organisms. In the laboratory, plasmids may be introduced into a cell via transformation. Synthetic plasmids are available for procurement over the in ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Chromosome
A chromosome is a long DNA molecule with part or all of the genetic material of an organism. In most chromosomes the very long thin DNA fibers are coated with packaging proteins; in eukaryotic cells the most important of these proteins are the histones. These proteins, aided by chaperone proteins, bind to and condense the DNA molecule to maintain its integrity. These chromosomes display a complex three-dimensional structure, which plays a significant role in transcriptional regulation. Chromosomes are normally visible under a light microscope only during the metaphase of cell division (where all chromosomes are aligned in the center of the cell in their condensed form). Before this happens, each chromosome is duplicated (S phase), and both copies are joined by a centromere, resulting either in an X-shaped structure (pictured above), if the centromere is located equatorially, or a two-arm structure, if the centromere is located distally. The joined copies are now ca ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DNA Replication
In molecular biology, DNA replication is the biological process of producing two identical replicas of DNA from one original DNA molecule. DNA replication occurs in all living organisms acting as the most essential part for biological inheritance. This is essential for cell division during growth and repair of damaged tissues, while it also ensures that each of the new cells receives its own copy of the DNA. The cell possesses the distinctive property of division, which makes replication of DNA essential. DNA is made up of a double helix of two complementary strands. The double helix describes the appearance of a double-stranded DNA which is thus composed of two linear strands that run opposite to each other and twist together to form. During replication, these strands are separated. Each strand of the original DNA molecule then serves as a template for the production of its counterpart, a process referred to as semiconservative replication. As a result of semi-conservativ ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cell Nucleus
The cell nucleus (pl. nuclei; from Latin or , meaning ''kernel'' or ''seed'') is a membrane-bound organelle found in eukaryotic cells. Eukaryotic cells usually have a single nucleus, but a few cell types, such as mammalian red blood cells, have no nuclei, and a few others including osteoclasts have many. The main structures making up the nucleus are the nuclear envelope, a double membrane that encloses the entire organelle and isolates its contents from the cellular cytoplasm; and the nuclear matrix, a network within the nucleus that adds mechanical support. The cell nucleus contains nearly all of the cell's genome. Nuclear DNA is often organized into multiple chromosomes – long stands of DNA dotted with various proteins, such as histones, that protect and organize the DNA. The genes within these chromosomes are structured in such a way to promote cell function. The nucleus maintains the integrity of genes and controls the activities of the cell by regulating g ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Histones
In biology, histones are highly basic proteins abundant in lysine and arginine residues that are found in eukaryotic cell nuclei. They act as spools around which DNA winds to create structural units called nucleosomes. Nucleosomes in turn are wrapped into 30-nanometer fibers that form tightly packed chromatin. Histones prevent DNA from becoming tangled and protect it from DNA damage. In addition, histones play important roles in gene regulation and DNA replication. Without histones, unwound DNA in chromosomes would be very long. For example, each human cell has about 1.8 meters of DNA if completely stretched out; however, when wound about histones, this length is reduced to about 90 micrometers (0.09 mm) of 30 nm diameter chromatin fibers. There are five families of histones which are designated H1/H5 (linker histones), H2, H3, and H4 (core histones). The nucleosome core is formed of two H2A-H2B dimers and a H3-H4 tetramer. The tight wrapping of DNA around histone ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |