|
Inbred Strains
Inbred strains (also called inbred lines, or rarely for animals linear animals) are individuals of a particular species which are nearly identical to each other in genotype due to long inbreeding. A strain is inbred when it has undergone at least 20 generations of brother x sister or offspring x parent mating, at which point at least 98.6% of the loci in an individual of the strain will be homozygous, and each individual can be treated effectively as clones. Some inbred strains have been bred for over 150 generations, leaving individuals in the population to be isogenic in nature. Inbred strains of animals are frequently used in laboratories for experiments where for the reproducibility of conclusions all the test animals should be as similar as possible. However, for some experiments, genetic diversity in the test population may be desired. Thus outbred strains of most laboratory animals are also available, where an outbred strain is a strain of an organism that is effectively wi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Species
In biology, a species is the basic unit of classification and a taxonomic rank of an organism, as well as a unit of biodiversity. A species is often defined as the largest group of organisms in which any two individuals of the appropriate sexes or mating types can produce fertile offspring, typically by sexual reproduction. Other ways of defining species include their karyotype, DNA sequence, morphology, behaviour or ecological niche. In addition, paleontologists use the concept of the chronospecies since fossil reproduction cannot be examined. The most recent rigorous estimate for the total number of species of eukaryotes is between 8 and 8.7 million. However, only about 14% of these had been described by 2011. All species (except viruses) are given a two-part name, a "binomial". The first part of a binomial is the genus to which the species belongs. The second part is called the specific name or the specific epithet (in botanical nomenclature, also sometimes i ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Monoclonal Antibodies
A monoclonal antibody (mAb, more rarely called moAb) is an antibody produced from a cell Lineage made by cloning a unique white blood cell. All subsequent antibodies derived this way trace back to a unique parent cell. Monoclonal antibodies can have monovalent affinity, binding only to the same epitope (the part of an antigen that is recognized by the antibody). In contrast, polyclonal antibodies bind to multiple epitopes and are usually made by several different antibody-secreting plasma cell lineages. Bispecific monoclonal antibodies can also be engineered, by increasing the therapeutic targets of one monoclonal antibody to two epitopes. It is possible to produce monoclonal antibodies that specifically bind to virtually any suitable substance; they can then serve to detect or purify it. This capability has become an investigative tool in biochemistry, molecular biology, and medicine. Monoclonal antibodies are being used on a clinical level for both the diagnosis and therapy ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Genetic Drift
Genetic drift, also known as allelic drift or the Wright effect, is the change in the frequency of an existing gene variant (allele) in a population due to random chance. Genetic drift may cause gene variants to disappear completely and thereby reduce genetic variation. It can also cause initially rare alleles to become much more frequent and even fixed. When few copies of an allele exist, the effect of genetic drift is more notable, and when many copies exist, the effect is less notable. In the middle of the 20th century, vigorous debates occurred over the relative importance of natural selection versus neutral processes, including genetic drift. Ronald Fisher, who explained natural selection using Mendelian genetics, held the view that genetic drift plays at most a minor role in evolution, and this remained the dominant view for several decades. In 1968, population geneticist Motoo Kimura rekindled the debate with his neutral theory of molecular evolution, which claims that ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Green Fluorescent Protein
The green fluorescent protein (GFP) is a protein that exhibits bright green fluorescence when exposed to light in the blue to ultraviolet range. The label ''GFP'' traditionally refers to the protein first isolated from the jellyfish ''Aequorea victoria'' and is sometimes called ''avGFP''. However, GFPs have been found in other organisms including corals, sea anemones, zoanithids, copepods and lancelets. The GFP from ''A. victoria'' has a major excitation peak at a wavelength of 395 nm and a minor one at 475 nm. Its emission peak is at 509 nm, which is in the lower green portion of the visible spectrum. The fluorescence quantum yield (QY) of GFP is 0.79. The GFP from the sea pansy (''Renilla reniformis'') has a single major excitation peak at 498 nm. GFP makes for an excellent tool in many forms of biology due to its ability to form an internal chromophore without requiring any accessory cofactors, gene products, or enzymes / substrates other than mo ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Genome
In the fields of molecular biology and genetics, a genome is all the genetic information of an organism. It consists of nucleotide sequences of DNA (or RNA in RNA viruses). The nuclear genome includes protein-coding genes and non-coding genes, other functional regions of the genome such as regulatory sequences (see non-coding DNA), and often a substantial fraction of 'junk' DNA with no evident function. Almost all eukaryotes have mitochondria and a small mitochondrial genome. Algae and plants also contain chloroplasts with a chloroplast genome. The study of the genome is called genomics. The genomes of many organisms have been sequenced and various regions have been annotated. The International Human Genome Project reported the sequence of the genome for ''Homo sapiens'' in 200The Human Genome Project although the initial "finished" sequence was missing 8% of the genome consisting mostly of repetitive sequences. With advancements in technology that could handle sequenci ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
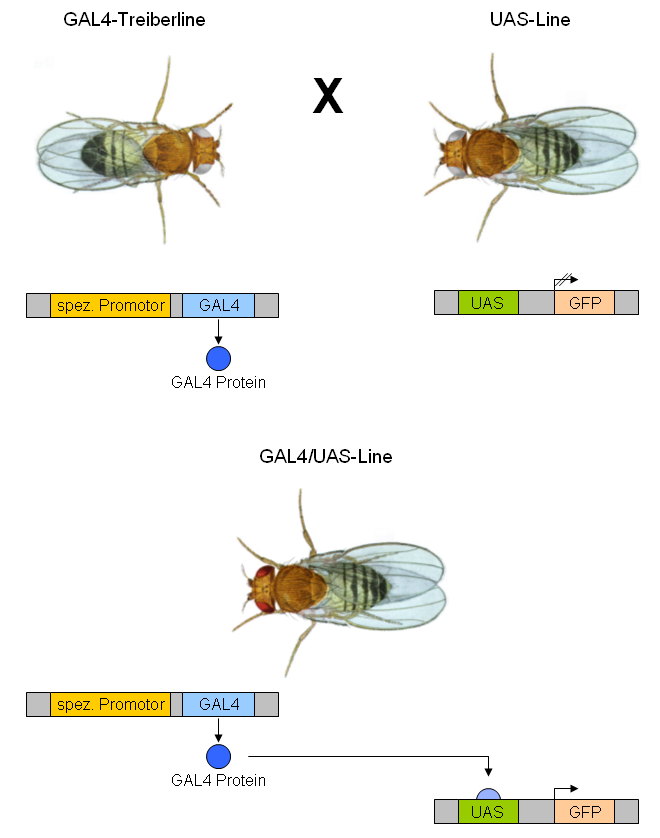
GAL4/UAS System
The GAL4-UAS system is a biochemical method used to study gene expression and function in organisms such as the fruit fly. It is based on the finding by Hitoshi Kakidani and Mark Ptashne, and Nicholas Webster and Pierre Chambon in 1988 that Gal4 binding to UAS sequences activates gene expression. The method was introduced into flies by Andrea Brand and Norbert Perrimon in 1993 and is considered a powerful technique for studying the expression of genes. The system has two parts: the Gal4 gene, encoding the yeast transcription activator protein Gal4, and the UAS (Upstream Activation Sequence), an enhancer to which GAL4 specifically binds to activate gene transcription. Overview The Gal4 system allows separation of the problems of defining which cells express a gene or protein and what the experimenter wants to do with this knowledge. Geneticists have created genetic variants of model organisms (typically fruit flies), called ''GAL4 lines'', each of which expresses GAL4 in some su ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Recombinant Inbred Strain
A recombinant inbred strain or recombinant inbred line (RIL) is an organism with chromosomes that incorporate an essentially permanent set of recombination events between chromosomes inherited from two or more inbred strains. F1 and F2 generations are produced by intercrossing the inbred strains; pairs of the F2 progeny are then mated to establish inbred strains through long-term inbreeding. Families of recombinant inbred strains numbering from 25 to 5000 are often used to map the locations of DNA sequence differences (quantitative trait loci) that contributed to differences in phenotype in model organisms. Recombinant inbred strains or lines were first developed using inbred strains of mice but are now used to study a wide range of organisms – ''Saccharomyces cerevisiae'' (yeast), ''Zea mays'' (maize), barley, ''Drosophila melanogaster'', ''C. elegans'' and rat. History The origins and history of recombinant inbred strains are described by Crow. While the potential utility of r ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Quantitative Trait Locus
A quantitative trait locus (QTL) is a locus (section of DNA) that correlates with variation of a quantitative trait in the phenotype of a population of organisms. QTLs are mapped by identifying which molecular markers (such as SNPs or AFLPs) correlate with an observed trait. This is often an early step in identifying the actual genes that cause the trait variation. Definition A quantitative trait locus (QTL) is a region of DNA which is associated with a particular phenotypic trait, which varies in degree and which can be attributed to polygenic effects, i.e., the product of two or more genes, and their environment. . These QTLs are often found on different chromosomes. The number of QTLs which explain variation in the phenotypic trait indicates the genetic architecture of a trait. It may indicate that plant height is controlled by many genes of small effect, or by a few genes of large effect. Typically, QTLs underlie continuous traits (those traits which vary continuously, ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
QTL Mapping Using Inbred Strains
A quantitative trait locus (QTL) is a locus (section of DNA) that correlates with variation of a quantitative trait in the phenotype of a population of organisms. QTLs are mapped by identifying which molecular markers (such as SNPs or AFLPs) correlate with an observed trait. This is often an early step in identifying the actual genes that cause the trait variation. Definition A quantitative trait locus (QTL) is a region of DNA which is associated with a particular phenotypic trait, which varies in degree and which can be attributed to polygenic effects, i.e., the product of two or more genes, and their environment. . These QTLs are often found on different chromosomes. The number of QTLs which explain variation in the phenotypic trait indicates the genetic architecture of a trait. It may indicate that plant height is controlled by many genes of small effect, or by a few genes of large effect. Typically, QTLs underlie continuous traits (those traits which vary continuously, ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Coisogenic Strain
Coisogenic strains are one type of inbred strain that differs by a mutation at a single locus and all of the other loci are identical. There are numerous ways to create an inbred strain and each of these strains are unique. Genetically engineered mice can be considered a coisogenic strain if the only difference between the engineered mouse and a wild-type mouse is a specific locus. Coisogenic strains can be used to investigate the function of a certain genetic locus. Coisogenic strains can be induced chemically or through radiation however, other types of alterations within the genome may also occur. Coisogenic strains may also occur through a spontaneous mutation that occurs in an inbred strain. To create a coisogenic strain through breeding, a mouse with the specific mutation on a locus is mated to an inbred strain (e.g., C57BL/6 C57BL/6, often referred to as "C57 black 6", "C57" or "black 6", is a common inbred strain of laboratory mouse. It is the most widely used "genet ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
FlyBase
FlyBase is an online bioinformatics database and the primary repository of genetic and molecular data for the insect family Drosophilidae. For the most extensively studied species and model organism, ''Drosophila melanogaster'', a wide range of data are presented in different formats. Information in FlyBase originates from a variety of sources ranging from large-scale genome projects to the primary research literature. These data types include mutant phenotypes; molecular characterization of mutant alleles; and other deviations, cytological maps, wild-type expression patterns, anatomical images, transgenic constructs and insertions, sequence-level gene models, and molecular classification of gene product functions. Query tools allow navigation of FlyBase through DNA or protein sequence, by gene or mutant name, or through terms from the several ontologies used to capture functional, phenotypic, and anatomical data. The database offers several different query tools in order to prov ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Jackson Laboratory
The Jackson Laboratory (often abbreviated as JAX) is an independent, non-profit biomedical research institution which was founded by a eugenicist. It employs more than 3,000 employees in Bar Harbor, Maine; Sacramento, California; Farmington, Connecticut; Shanghai, China; and Yokohama, Japan. The institution is a National Cancer Institute-designated Cancer Center and has NIH Centers of Excellence in aging and systems genetics. The mission of The Jackson Laboratory is "to discover the genetic basis for preventing, treating and curing human diseases, and to enable research and education for the global biomedical community." The laboratory is also the world's source for more than 8,000 strains of genetically defined mice, home of the Mouse Genome Informatics database, and is an international hub for scientific courses, conferences, training and education. Major research areas Jackson Laboratory's research, represented by the activities of more than 60 laboratories, performs resear ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |