|
Heteronuclear Single Quantum Coherence
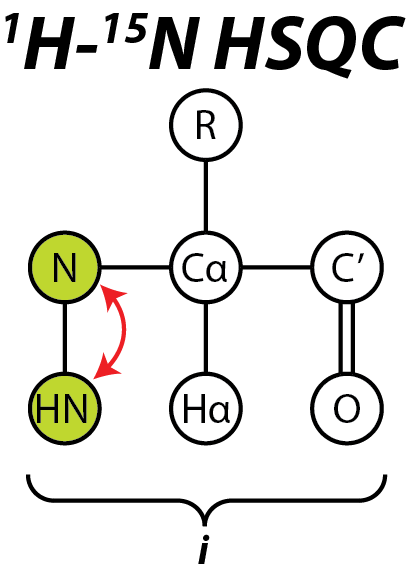
The heteronuclear single quantum coherence or heteronuclear single quantum correlation experiment, normally abbreviated as HSQC, is used frequently in NMR spectroscopy of organic molecules and is of particular significance in the field of protein NMR. The experiment was first described by Geoffrey Bodenhausen and D. J. Ruben in 1980. The resulting spectrum is two-dimensional (2D) with one axis for proton (1H) and the other for a heteronucleus (an atomic nucleus other than a proton), which is usually 13C or 15N. The spectrum contains a peak for each unique proton attached to the heteronucleus being considered. The 2D HSQC can also be combined with other experiments in higher-dimensional NMR experiments, such as NOESY-HSQC or TOCSY-HSQC. General scheme The HSQC experiment is a highly sensitive 2D-NMR experiment and was first described in a 1H—15N system, but is also applicable to other nuclei such as 1H—13C and 1H—31P. The basic scheme of this experiment involves the tran ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein NMR
Nuclear magnetic resonance spectroscopy of proteins (usually abbreviated protein NMR) is a field of structural biology in which NMR spectroscopy is used to obtain information about the structure and dynamics of proteins, and also nucleic acids, and their complexes. The field was pioneered by Richard R. Ernst and Kurt Wüthrich at the ETH, and by Ad Bax, Marius Clore, Angela Gronenborn at the NIH, and Gerhard Wagner at Harvard University, among others. Structure determination by NMR spectroscopy usually consists of several phases, each using a separate set of highly specialized techniques. The sample is prepared, measurements are made, interpretive approaches are applied, and a structure is calculated and validated. NMR involves the quantum-mechanical properties of the central core ("nucleus") of the atom. These properties depend on the local molecular environment, and their measurement provides a map of how the atoms are linked chemically, how close they are in space, and how rapid ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Asparagine
Asparagine (symbol Asn or N) is an α-amino acid that is used in the biosynthesis of proteins. It contains an α-amino group (which is in the protonated −NH form under biological conditions), an α-carboxylic acid group (which is in the deprotonated −COO− form under biological conditions), and a side chain carboxamide, classifying it as a polar (at physiological pH), aliphatic amino acid. It is non-essential in humans, meaning the body can synthesize it. It is encoded by the codons AAU and AAC. History Asparagine was first isolated in 1806 in a crystalline form by French chemists Louis Nicolas Vauquelin and Pierre Jean Robiquet (then a young assistant). It was isolated from asparagus juice, in which it is abundant, hence the chosen name. It was the first amino acid to be isolated. Three years later, in 1809, Pierre Jean Robiquet identified a substance from liquorice root with properties which he qualified as very similar to those of asparagine, and which Plisson identi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein NMR
Nuclear magnetic resonance spectroscopy of proteins (usually abbreviated protein NMR) is a field of structural biology in which NMR spectroscopy is used to obtain information about the structure and dynamics of proteins, and also nucleic acids, and their complexes. The field was pioneered by Richard R. Ernst and Kurt Wüthrich at the ETH, and by Ad Bax, Marius Clore, Angela Gronenborn at the NIH, and Gerhard Wagner at Harvard University, among others. Structure determination by NMR spectroscopy usually consists of several phases, each using a separate set of highly specialized techniques. The sample is prepared, measurements are made, interpretive approaches are applied, and a structure is calculated and validated. NMR involves the quantum-mechanical properties of the central core ("nucleus") of the atom. These properties depend on the local molecular environment, and their measurement provides a map of how the atoms are linked chemically, how close they are in space, and how rapid ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Leucine
Leucine (symbol Leu or L) is an essential amino acid that is used in the biosynthesis of proteins. Leucine is an α-amino acid, meaning it contains an α-amino group (which is in the protonated −NH3+ form under biological conditions), an α- carboxylic acid group (which is in the deprotonated −COO− form under biological conditions), and a side chain isobutyl group, making it a non-polar aliphatic amino acid. It is essential in humans, meaning the body cannot synthesize it: it must be obtained from the diet. Human dietary sources are foods that contain protein, such as meats, dairy products, soy products, and beans and other legumes. It is encoded by the codons UUA, UUG, CUU, CUC, CUA, and CUG. Like valine and isoleucine, leucine is a branched-chain amino acid. The primary metabolic end products of leucine metabolism are acetyl-CoA and acetoacetate; consequently, it is one of the two exclusively ketogenic amino acids, with lysine being the other. It is the most import ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Alpha And Beta Carbon
In the nomenclature of organic chemistry, a locant is a term to indicate the position of a functional group or substituent within a molecule. Numeric locants The International Union of Pure and Applied Chemistry (IUPAC) recommends the use of numeric prefixes to indicate the position of substituents, generally by identifying the parent hydrocarbon chain and assigning the carbon atoms based on their substituents in order of precedence. For example, there are at least two isomers of the linear form of pentanone, a ketone that contains a chain of exactly five carbon atoms. There is an oxygen atom bonded to one of the middle three carbons (if it were bonded to an end carbon, the molecule would be an aldehyde, not a ketone), but it is not clear where it is located. In this example, the carbon atoms are numbered from one to five, which starts at one end and proceeds sequentially along the chain. Now the position of the oxygen atom can be defined as on carbon atom number two, three or ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
J-coupling
In nuclear chemistry and nuclear physics, ''J''-couplings (also called spin-spin coupling or indirect dipole–dipole coupling) are mediated through chemical bonds connecting two spins. It is an indirect interaction between two nuclear spins that arises from hyperfine interactions between the nuclei and local electrons. In NMR spectroscopy, ''J''-coupling contains information about relative bond distances and angles. Most importantly, ''J''-coupling provides information on the connectivity of chemical bonds. It is responsible for the often complex splitting of resonance lines in the NMR spectra of fairly simple molecules. ''J''-coupling is a frequency ''difference'' that is not affected by the strength of the magnetic field, so is always stated in Hz. Vector model and manifestations for chemical structure assignments The origin of ''J''-coupling can be visualized by a vector model for a simple molecule such as hydrogen fluoride (HF). In HF, the two nuclei have spin . Four states ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ionization Constant
In chemistry, an acid dissociation constant (also known as acidity constant, or acid-ionization constant; denoted ) is a quantitative measure of the strength of an acid in solution. It is the equilibrium constant for a chemical reaction :HA A^- + H^+ known as dissociation in the context of acid–base reactions. The chemical species HA is an acid that dissociates into , the conjugate base of the acid and a hydrogen ion, . The system is said to be in equilibrium when the concentrations of its components will not change over time, because both forward and backward reactions are occurring at the same rate. The dissociation constant is defined by :K_\text = \mathrm, or :\mathrmK_\ce = - \log_ K_\text = \log_\frac where quantities in square brackets represent the concentrations of the species at equilibrium. Theoretical background The acid dissociation constant for an acid is a direct consequence of the underlying thermodynamics of the dissociation reaction; the p''K''a valu ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ligand
In coordination chemistry, a ligand is an ion or molecule (functional group) that binds to a central metal atom to form a coordination complex. The bonding with the metal generally involves formal donation of one or more of the ligand's electron pairs, often through Lewis bases. The nature of metal–ligand bonding can range from covalent to ionic. Furthermore, the metal–ligand bond order can range from one to three. Ligands are viewed as Lewis bases, although rare cases are known to involve Lewis acidic "ligands". Metals and metalloids are bound to ligands in almost all circumstances, although gaseous "naked" metal ions can be generated in a high vacuum. Ligands in a complex dictate the reactivity of the central atom, including ligand substitution rates, the reactivity of the ligands themselves, and redox. Ligand selection requires critical consideration in many practical areas, including bioinorganic and medicinal chemistry, homogeneous catalysis, and environmental chemi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Structure
Protein structure is the three-dimensional arrangement of atoms in an amino acid-chain molecule. Proteins are polymers specifically polypeptides formed from sequences of amino acids, the monomers of the polymer. A single amino acid monomer may also be called a ''residue'' indicating a repeating unit of a polymer. Proteins form by amino acids undergoing condensation reactions, in which the amino acids lose one water molecule per reaction in order to attach to one another with a peptide bond. By convention, a chain under 30 amino acids is often identified as a peptide, rather than a protein. To be able to perform their biological function, proteins fold into one or more specific spatial conformations driven by a number of non-covalent interactions such as hydrogen bonding, ionic interactions, Van der Waals forces, and hydrophobic packing. To understand the functions of proteins at a molecular level, it is often necessary to determine their three-dimensional structure. This is t ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Relaxation (NMR)
In MRI and NMR spectroscopy, an observable nuclear spin polarization ( magnetization) is created by a homogeneous magnetic field. This field makes the magnetic dipole moments of the sample precess at the resonance ( Larmor) frequency of the nuclei. At thermal equilibrium, nuclear spins precess randomly about the direction of the applied field. They become abruptly phase coherent when they are hit by radiofrequent (RF) pulses at the resonant frequency, created orthogonal to the field. The RF pulses cause the population of spin-states to be perturbed from their thermal equilibrium value. The generated transverse magnetization can then induce a signal in an RF coil that can be detected and amplified by an RF receiver. The return of the longitudinal component of the magnetization to its equilibrium value is termed ''spin-lattice'' relaxation while the loss of phase-coherence of the spins is termed ''spin-spin'' relaxation, which is manifest as an observed free induction decay (FID). ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |




4-3D-balls.png)

