|
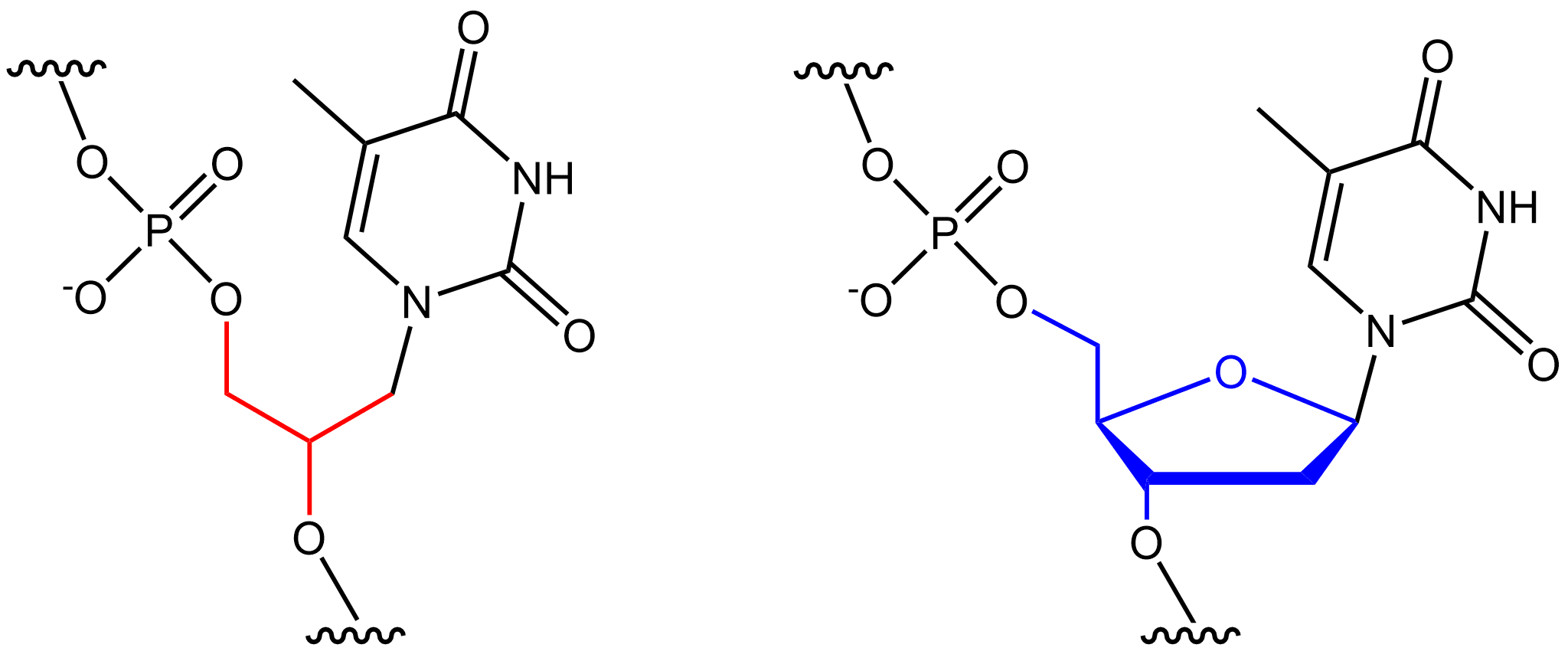
Glycol Nucleic Acid
Glycol nucleic acid (GNA), sometimes also referred to as glycerol nucleic acid, is a nucleic acid similar to DNA or RNA but differing in the composition of its sugar-phosphodiester backbone, using propylene glycol in place of ribose or deoxyribose. GNA is chemically stable but not known to occur naturally. However, due to its simplicity, it might have played a role in the evolution of life. The 2,3-dihydroxypropyl nucleoside analogues were first prepared by Ueda et al. (1971). Soon thereafter it was shown that phosphate-linked oligomers of the analogues do in fact exhibit hypochromicity in the presence of RNA and DNA in solution (Seita et al. 1972). The preparation of the polymers was later described by Cook et al. (1995, 1999) and Acevedo and Andrews (1996). However the ability of GNA-GNA self-pairing was first reported by Zhang and Meggers in 2005. Crystal structures of a GNA duplexes were subsequently reported by Essen and Meggers. DNA and RNA have a deoxyribose and ribose sug ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Propylene Glycol
Propylene glycol (IUPAC nomenclature, IUPAC name: propane-1,2-diol) is a viscous, colorless liquid, which is nearly odorless but possesses a faintly sweet taste. Its chemical formula is CH3CH(OH)CH2OH. Containing two Alcohol (chemistry), alcohol groups, it is classed as a diol. It is miscible with a broad range of solvents, including water, acetone, and chloroform. In general, glycols are non-irritating and have very low Volatility (chemistry), volatility. It is produced on a large scale primarily for the production of polymers. In the European Union, it has E-number E1520 for food applications. For cosmetics and pharmacology, the number is E490. Propylene glycol is also present in propylene glycol alginate, which is known as E405. Propylene glycol is a compound which is Generally recognized as safe, GRAS (generally recognized as safe) by the US Food and Drug Administration under 21 CFR x184.1666, and is also approved by the FDA for certain uses as an indirect food additive. Pro ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Oligomer
In chemistry and biochemistry, an oligomer () is a molecule that consists of a few repeating units which could be derived, actually or conceptually, from smaller molecules, monomers.Quote: ''Oligomer molecule: A molecule of intermediate relative molecular mass, the structure of which essentially comprises a small plurality of units derived, actually or conceptually, from molecules of lower relative molecular mass.'' The name is composed of Greek elements '' oligo-'', "a few" and '' -mer'', "parts". An adjective form is ''oligomeric''. The oligomer concept is contrasted to that of a polymer, which is usually understood to have a large number of units, possibly thousands or millions. However, there is no sharp distinction between these two concepts. One proposed criterion is whether the molecule's properties vary significantly with the removal of one or a few of the units. An oligomer with a specific number of units is referred to by the Greek prefix denoting that number, wi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Deoxyribose
Deoxyribose, or more precisely 2-deoxyribose, is a monosaccharide with idealized formula H−(C=O)−(CH2)−(CHOH)3−H. Its name indicates that it is a deoxy sugar, meaning that it is derived from the sugar ribose by loss of a hydroxy group. Discovered in 1929 by Phoebus Levene, deoxyribose is most notable for its presence in DNA. Since the pentose sugars arabinose and ribose only differ by the stereochemistry at C2′, 2-deoxyribose and 2-deoxyarabinose are equivalent, although the latter term is rarely used because ribose, not arabinose, is the precursor to deoxyribose. Structure Several isomers exist with the formula H−(C=O)−(CH2)−(CHOH)3−H, but in deoxyribose all the hydroxyl groups are on the same side in the Fischer projection. The term "2-deoxyribose" may refer to either of two enantiomers: the biologically important -2-deoxyribose and to the rarely encountered mirror image -2-deoxyribose.C Bernelot-Moens and B Demple (1989), ''Multiple DNA repair activities f ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ribose
Ribose is a simple sugar and carbohydrate with molecular formula C5H10O5 and the linear-form composition H−(C=O)−(CHOH)4−H. The naturally-occurring form, , is a component of the ribonucleotides from which RNA is built, and so this compound is necessary for coding, decoding, regulation and expression of genes. It has a structural analog, deoxyribose, which is a similarly essential component of DNA. is an unnatural sugar that was first prepared by Emil Fischer and Oscar Piloty in 1891. It was not until 1909 that Phoebus Levene and Walter Jacobs recognised that was a natural product, the enantiomer of Fischer and Piloty's product, and an essential component of nucleic acids. Fischer chose the name "ribose" as it is a partial rearrangement of the name of another sugar, arabinose, of which ribose is an epimer at the 2' carbon; both names also relate to gum arabic, from which arabinose was first isolated and from which they prepared . Like most sugars, ribose exists ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Phosphodiester Bond
In chemistry, a phosphodiester bond occurs when exactly two of the hydroxyl groups () in phosphoric acid react with hydroxyl groups on other molecules to form two ester bonds. The "bond" involves this linkage . Discussion of phosphodiesters is dominated by their prevalence in DNA and RNA, but phosphodiesters occur in other biomolecules, e.g. acyl carrier proteins. Phosphodiester bonds make up the backbones of DNA and RNA. The phosphate is attached to the 5' carbon. The 3' carbon of one sugar is bonded to the 5' phosphate of the adjacent sugar. Specifically, the phosphodiester bond links the 3' carbon atom of one sugar molecule and the 5' carbon atom of another (hence the name, 3', 5' phosphodiester linkage). These saccharide groups are derived from deoxyribose in DNA and ribose in RNA. Phosphodiesters are negatively charged at pH 7. Repulsion between these negative charges influences the conformation of the polynucleic acids. The negative charge attracts histones, metal c ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Carbon
Carbon () is a chemical element with the symbol C and atomic number 6. It is nonmetallic and tetravalent In chemistry, the valence (US spelling) or valency (British spelling) of an element is the measure of its combining capacity with other atoms when it forms chemical compounds or molecules. Description The combining capacity, or affinity of an ...—its atom making four electrons available to form covalent bond, covalent chemical bonds. It belongs to group 14 of the periodic table. Carbon makes up only about 0.025 percent of Earth's crust. Three Isotopes of carbon, isotopes occur naturally, Carbon-12, C and Carbon-13, C being stable, while Carbon-14, C is a radionuclide, decaying with a half-life of about 5,730 years. Carbon is one of the Timeline of chemical element discoveries#Ancient discoveries, few elements known since antiquity. Carbon is the 15th Abundance of elements in Earth's crust, most abundant element in the Earth's crust, and the Abundance of the c ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Watson-Crick Base Pairing
A base pair (bp) is a fundamental unit of double-stranded nucleic acids consisting of two nucleobases bound to each other by hydrogen bonds. They form the building blocks of the DNA double helix and contribute to the folded structure of both DNA and RNA. Dictated by specific hydrogen bonding patterns, "Watson–Crick" (or "Watson–Crick–Franklin") base pairs (guanine–cytosine and adenine–thymine) allow the DNA helix to maintain a regular helical structure that is subtly dependent on its nucleotide sequence. The complementary nature of this based-paired structure provides a redundant copy of the genetic information encoded within each strand of DNA. The regular structure and data redundancy provided by the DNA double helix make DNA well suited to the storage of genetic information, while base-pairing between DNA and incoming nucleotides provides the mechanism through which DNA polymerase replicates DNA and RNA polymerase transcribes DNA into RNA. Many DNA-binding protei ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Abiogenesis
In biology, abiogenesis (from a- 'not' + Greek bios 'life' + genesis 'origin') or the origin of life is the natural process by which life has arisen from non-living matter, such as simple organic compounds. The prevailing scientific hypothesis is that the transition from non-living to living entities on Earth was not a single event, but an evolutionary process of increasing complexity that involved the formation of a habitable planet, the prebiotic synthesis of organic molecules, molecular self-replication, self-assembly, autocatalysis, and the emergence of cell membranes. Many proposals have been made for different stages of the process. The study of abiogenesis aims to determine how pre-life chemical reactions gave rise to life under conditions strikingly different from those on Earth today. It primarily uses tools from biology and chemistry, with more recent approaches attempting a synthesis of many sciences. Life functions through the specialized chemistry of carbon and ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Locked Nucleic Acid
A locked nucleic acid (LNA), also known as bridged nucleic acid (BNA), and often referred to as inaccessible RNA, is a modified RNA nucleotide in which the ribose moiety is modified with an extra bridge connecting the 2' oxygen and 4' carbon. The bridge "locks" the ribose in the 3'-''endo'' (North) conformation, which is often found in the A-form duplexes. This structure provides for increased stability against enzymatic degradation. LNA also offers improved specificity and affinity in base-pairing as a monomer or a constituent of an oligonucleotide. LNA nucleotides can be mixed with DNA or RNA residues in a oligonucleotide. Synthesis Obika et al. were the first to chemically synthesize LNA in 1997, independently followed by Jesper Wengel's group in 1998. This became possible after Zamecnick and Stephenson laid the groundwork on the possibility of oligonucleotides being great agents for controlling gene expression in 1978. To date, two different approaches, referred to as linear ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Oligonucleotide Synthesis
Oligonucleotide synthesis is the chemical synthesis of relatively short fragments of nucleic acids with defined chemical structure (sequence). The technique is extremely useful in current laboratory practice because it provides a rapid and inexpensive access to custom-made oligonucleotides of the desired sequence. Whereas enzymes synthesize DNA and RNA only in a 5' to 3' direction, chemical oligonucleotide synthesis does not have this limitation, although it is most often carried out in the opposite, 3' to 5' direction. Currently, the process is implemented as solid-phase synthesis using phosphoramidite method and phosphoramidite building blocks derived from protected 2'-deoxynucleosides ( dA, dC, dG, and T), ribonucleosides ( A, C, G, and U), or chemically modified nucleosides, e.g. LNA or BNA. To obtain the desired oligonucleotide, the building blocks are sequentially coupled to the growing oligonucleotide chain in the order required by the sequence of the product (see ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Peptide Nucleic Acid
Peptide nucleic acid (PNA) is an artificially synthesized polymer similar to DNA or RNA. Synthetic peptide nucleic acid oligomers have been used in recent years in molecular biology procedures, diagnostic assays, and antisense therapies. Due to their higher binding strength, it is not necessary to design long PNA oligomers for use in these roles, which usually require oligonucleotide probes of 20–25 bases. The main concern of the length of the PNA-oligomers is to guarantee the specificity. PNA oligomers also show greater specificity in binding to complementary DNAs, with a PNA/DNA base mismatch being more destabilizing than a similar mismatch in a DNA/DNA duplex. This binding strength and specificity also applies to PNA/RNA duplexes. PNAs are not easily recognized by either nucleases or proteases, making them resistant to degradation by enzymes. PNAs are also stable over a wide pH range. Though an unmodified PNA cannot readily cross the cell membrane to enter the cytosol, cov ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |