|
Glutamate Permease
The sodium/glutamate symporter, also known as glutamate permease, is a transmembrane protein family found in bacteria and archaea. These proteins are symporters that are responsible for the sodium-dependent uptake of extracellular glutamate into the cell. They are integral membrane proteins located in the bacterial inner membrane. The best-studied member of the family is GltS from ''Escherichia coli''. GltS contains ten transmembrane helices arranged in two antiparallel 5-helix domains and functions as a homodimer. Substrates for GltS include L- and D-glutamate, as well as toxic analogs α- methylglutamate, and homocysteate. In studies of ''E. coli'' growth, bacteria without GltS were unable to grow in a medium where glutamate is the only source of carbon. The family is evolutionarily distant from other glutamate transporters. Phylogenetic analyses of GltS genes suggest that their presence in cyanobacteria Cyanobacteria (), also known as Cyanophyta, are a phylum of gram- ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
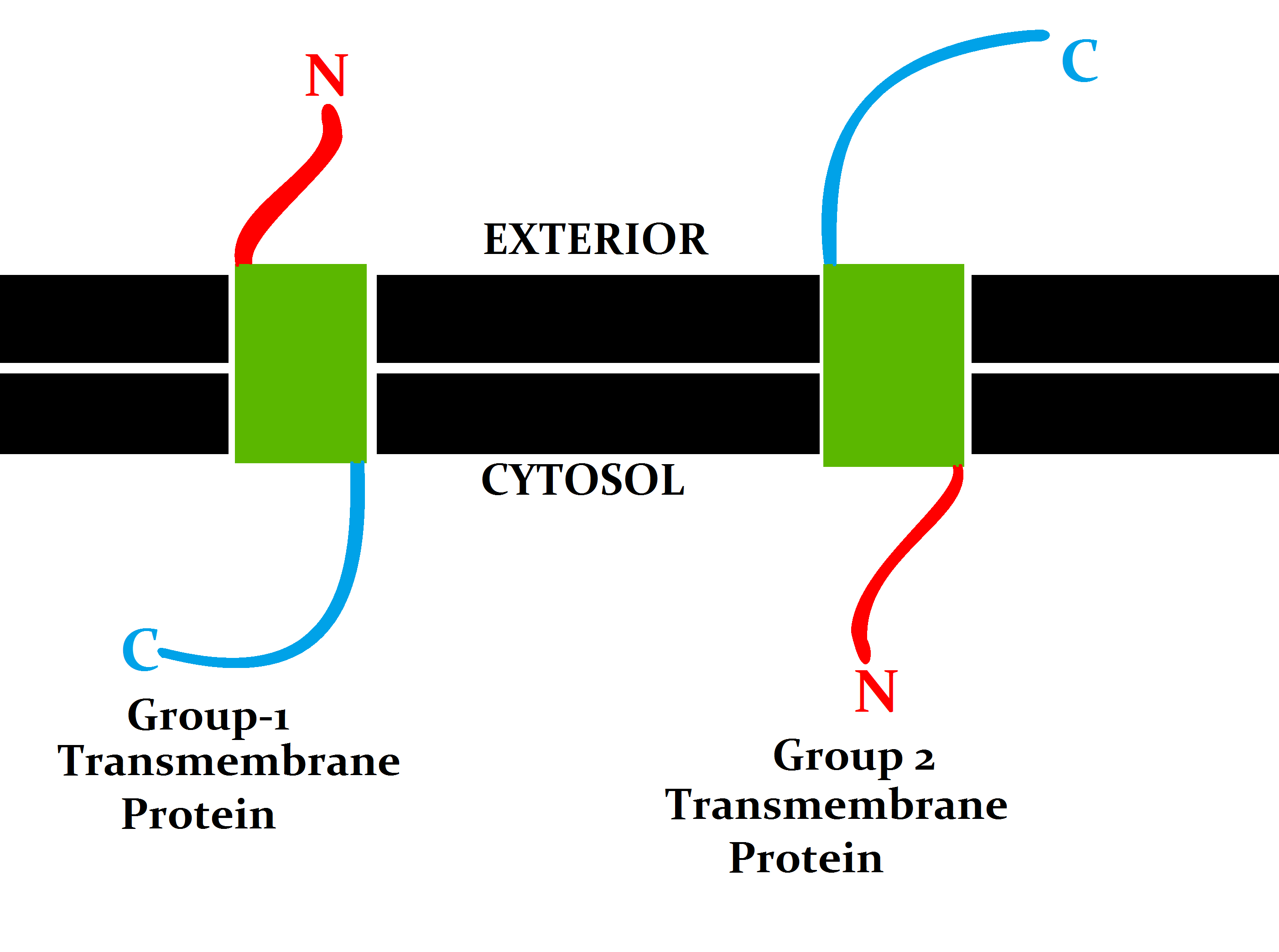
Transmembrane Protein
A transmembrane protein (TP) is a type of integral membrane protein that spans the entirety of the cell membrane. Many transmembrane proteins function as gateways to permit the transport of specific substances across the membrane. They frequently undergo significant conformational changes to move a substance through the membrane. They are usually highly hydrophobic and aggregate and precipitate in water. They require detergents or nonpolar solvents for extraction, although some of them (beta-barrels) can be also extracted using denaturing agents. The peptide sequence that spans the membrane, or the transmembrane segment, is largely hydrophobic and can be visualized using the hydropathy plot. Depending on the number of transmembrane segments, transmembrane proteins can be classified as single-span (or bitopic) or multi-span (polytopic). Some other integral membrane proteins are called monotopic, meaning that they are also permanently attached to the membrane, but do not pass ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Transmembrane Helices
A transmembrane domain (TMD) is a membrane-spanning protein domain. TMDs generally adopt an alpha helix topological conformation, although some TMDs such as those in porins can adopt a different conformation. Because the interior of the lipid bilayer is hydrophobic, the amino acid residues in TMDs are often hydrophobic, although proteins such as membrane pumps and ion channels can contain polar residues. TMDs vary greatly in length, sequence, and hydrophobicity, adopting organelle-specific properties. Functions of transmembrane domains Transmembrane domains are known to perform a variety of functions. These include: * Anchoring transmembrane proteins to the membrane. *Facilitating molecular transport of molecules such as ions and proteins across biological membranes; usually hydrophilic residues and binding sites in the TMDs help in this process. *Signal transduction across the membrane; many transmembrane proteins, such as G protein-coupled receptors, receive extracellular s ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cyanobacteria
Cyanobacteria (), also known as Cyanophyta, are a phylum of gram-negative bacteria that obtain energy via photosynthesis. The name ''cyanobacteria'' refers to their color (), which similarly forms the basis of cyanobacteria's common name, blue-green algae, although they are not usually scientifically classified as algae. They appear to have originated in a freshwater or terrestrial environment. Sericytochromatia, the proposed name of the paraphyletic and most basal group, is the ancestor of both the non-photosynthetic group Melainabacteria and the photosynthetic cyanobacteria, also called Oxyphotobacteria. Cyanobacteria use photosynthetic pigments, such as carotenoids, phycobilins, and various forms of chlorophyll, which absorb energy from light. Unlike heterotrophic prokaryotes, cyanobacteria have internal membranes. These are flattened sacs called thylakoids where photosynthesis is performed. Phototrophic eukaryotes such as green plants perform photosynthesis in plast ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Gene
In biology, the word gene (from , ; "...Wilhelm Johannsen coined the word gene to describe the Mendelian units of heredity..." meaning ''generation'' or ''birth'' or ''gender'') can have several different meanings. The Mendelian gene is a basic unit of heredity and the molecular gene is a sequence of nucleotides in DNA that is transcribed to produce a functional RNA. There are two types of molecular genes: protein-coding genes and noncoding genes. During gene expression, the DNA is first copied into RNA. The RNA can be directly functional or be the intermediate template for a protein that performs a function. The transmission of genes to an organism's offspring is the basis of the inheritance of phenotypic traits. These genes make up different DNA sequences called genotypes. Genotypes along with environmental and developmental factors determine what the phenotypes will be. Most biological traits are under the influence of polygenes (many different genes) as well as gen ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Phylogenetic
In biology, phylogenetics (; from Greek φυλή/ φῦλον [] "tribe, clan, race", and wikt:γενετικός, γενετικός [] "origin, source, birth") is the study of the evolutionary history and relationships among or within groups of organisms. These relationships are determined by Computational phylogenetics, phylogenetic inference methods that focus on observed heritable traits, such as DNA sequences, protein amino acid sequences, or morphology. The result of such an analysis is a phylogenetic tree—a diagram containing a hypothesis of relationships that reflects the evolutionary history of a group of organisms. The tips of a phylogenetic tree can be living taxa or fossils, and represent the "end" or the present time in an evolutionary lineage. A phylogenetic diagram can be rooted or unrooted. A rooted tree diagram indicates the hypothetical common ancestor of the tree. An unrooted tree diagram (a network) makes no assumption about the ancestral line, and does ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Homocysteate
Homocysteic acid is a sulfur-containing glutamic acid analog and a potent NMDA receptor agonist. It is related to homocysteine, a by-product of methionine Methionine (symbol Met or M) () is an essential amino acid in humans. As the precursor of other amino acids such as cysteine and taurine, versatile compounds such as SAM-e, and the important antioxidant glutathione, methionine plays a critical ro ... metabolism. References Amino acids Sulfonic acids {{organic-compound-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Methylglutamate
''N''-Methyl--glutamic acid (methylglutamate) is a chemical derivative of glutamic acid in which a methyl group has been added to the amino group. It is an intermediate in methane metabolism. Biosynthetically, it is produced from methylamine and glutamic acid Glutamic acid (symbol Glu or E; the ionic form is known as glutamate) is an α-amino acid that is used by almost all living beings in the biosynthesis of proteins. It is a non-essential nutrient for humans, meaning that the human body can synt ... by the enzyme methylamine—glutamate ''N''-methyltransferase. It can also be demethylated by methylglutamate dehydrogenase to regenerate glutamic acid. References Amino acid derivatives Dicarboxylic acids Secondary amino acids {{organic-compound-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
D Amino Acid
D-Amino acids are amino acids where the stereogenic carbon alpha to the amino group has the D-configuration. For most naturally-occurring amino acids, this carbon has the L-configuration. D-Amino acids are occasionally found in nature as residues in proteins. They are formed from ribosomally-derived D-amino acid residues. Amino acids, as components of peptides, peptide hormones, structural and immune proteins, are the most important bioregulators involved in all life processes along with nucleic acids, carbohydrates and lipids. "Environmental D-amino acids are thought to be derived from organic diagenesis such as racemization and release from bacterial cell walls and even from microbial production." Discovery Their discovery was in the 1950s. "Auclair and Patton (1950) first reported their presence in the blood of insects and mollusks" Furthermore, they also have been identified in various mammalian tissues. The two major types of D-amino acids synthesized in and by mam ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Homodimer
In biochemistry, a protein dimer is a macromolecular complex formed by two protein monomers, or single proteins, which are usually non-covalently bound. Many macromolecules, such as proteins or nucleic acids, form dimers. The word ''dimer'' has roots meaning "two parts", '' di-'' + '' -mer''. A protein dimer is a type of protein quaternary structure. A protein homodimer is formed by two identical proteins. A protein heterodimer is formed by two different proteins. Most protein dimers in biochemistry are not connected by covalent bonds. An example of a non-covalent heterodimer is the enzyme reverse transcriptase, which is composed of two different amino acid chains. An exception is dimers that are linked by disulfide bridges such as the homodimeric protein NEMO. Some proteins contain specialized domains to ensure dimerization (dimerization domains) and specificity. The G protein-coupled cannabinoid receptors have the ability to form both homo- and heterodimers with several ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Escherichia Coli
''Escherichia coli'' (),Wells, J. C. (2000) Longman Pronunciation Dictionary. Harlow ngland Pearson Education Ltd. also known as ''E. coli'' (), is a Gram-negative, facultative anaerobic, rod-shaped, coliform bacterium of the genus ''Escherichia'' that is commonly found in the lower intestine of warm-blooded organisms. Most ''E. coli'' strains are harmless, but some serotypes ( EPEC, ETEC etc.) can cause serious food poisoning in their hosts, and are occasionally responsible for food contamination incidents that prompt product recalls. Most strains do not cause disease in humans and are part of the normal microbiota of the gut; such strains are harmless or even beneficial to humans (although these strains tend to be less studied than the pathogenic ones). For example, some strains of ''E. coli'' benefit their hosts by producing vitamin K2 or by preventing the colonization of the intestine by pathogenic bacteria. These mutually beneficial relationships between ''E. col ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Family
A protein family is a group of evolutionarily related proteins. In many cases, a protein family has a corresponding gene family, in which each gene encodes a corresponding protein with a 1:1 relationship. The term "protein family" should not be confused with Family (biology), family as it is used in taxonomy. Proteins in a family descend from a common ancestor and typically have similar protein structure, three-dimensional structures, functions, and significant Sequence homology, sequence similarity. The most important of these is sequence similarity (usually amino-acid sequence), since it is the strictest indicator of homology and therefore the clearest indicator of common ancestry. A fairly well developed framework exists for evaluating the significance of similarity between a group of sequences using sequence alignment methods. Proteins that do not share a common ancestor are very unlikely to show statistically significant sequence similarity, making sequence alignment a powerf ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Bacterial Inner Membrane
Bacteria (; singular: bacterium) are ubiquitous, mostly free-living organisms often consisting of one biological cell. They constitute a large domain of prokaryotic microorganisms. Typically a few micrometres in length, bacteria were among the first life forms to appear on Earth, and are present in most of its habitats. Bacteria inhabit soil, water, acidic hot springs, radioactive waste, and the deep biosphere of Earth's crust. Bacteria are vital in many stages of the nutrient cycle by recycling nutrients such as the fixation of nitrogen from the atmosphere. The nutrient cycle includes the decomposition of dead bodies; bacteria are responsible for the putrefaction stage in this process. In the biological communities surrounding hydrothermal vents and cold seeps, extremophile bacteria provide the nutrients needed to sustain life by converting dissolved compounds, such as hydrogen sulphide and methane, to energy. Bacteria also live in symbiotic and parasitic relationships wi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
.png)






