|
Gb1 Domain
Protein G is an immunoglobulin-binding protein expressed in group C and G Streptococcal bacteria much like Protein A but with differing binding specificities. It is a ~60-kDA (65 kDA for strain G148 and 58 kDa for strain C40) cell surface protein that has found application in purifying antibodies through its binding to the Fab and Fc region. The native molecule also binds albumin, but because serum albumin is a major contaminant of antibody sources, the albumin binding site has been removed from recombinant forms of Protein G. This recombinant Protein G, either labeled with a fluorophore or a single-stranded DNA strand, was used as a replacement for secondary antibodies in immunofluorescence and super-resolution imaging. Other antibody binding proteins In addition to Protein G, other immunoglobulin-binding bacterial proteins such as Protein A, Protein A/G and Protein L are all commonly used to purify, immobilize or detect immunoglobulins. Each of these immunoglobulin-binding ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Immunoglobulin
An antibody (Ab), also known as an immunoglobulin (Ig), is a large, Y-shaped protein used by the immune system to identify and neutralize foreign objects such as pathogenic bacteria and viruses. The antibody recognizes a unique molecule of the pathogen, called an antigen. Each tip of the "Y" of an antibody contains a paratope (analogous to a lock) that is specific for one particular epitope (analogous to a key) on an antigen, allowing these two structures to bind together with precision. Using this binding mechanism, an antibody can ''tag'' a microbe or an infected cell for attack by other parts of the immune system, or can neutralize it directly (for example, by blocking a part of a virus that is essential for its invasion). To allow the immune system to recognize millions of different antigens, the antigen-binding sites at both tips of the antibody come in an equally wide variety. In contrast, the remainder of the antibody is relatively constant. It only occurs in a few vari ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Nucleation
In thermodynamics, nucleation is the first step in the formation of either a new thermodynamic phase or structure via self-assembly or self-organization within a substance or mixture. Nucleation is typically defined to be the process that determines how long an observer has to wait before the new phase or self-organized structure appears. For example, if a volume of water is cooled (at atmospheric pressure) below 0°C, it will tend to freeze into ice, but volumes of water cooled only a few degrees below 0°C often stay completely free of ice for long periods (supercooling). At these conditions, nucleation of ice is either slow or does not occur at all. However, at lower temperatures nucleation is fast, and ice crystals appear after little or no delay. Nucleation is a common mechanism which generates first-order phase transitions, and it is the start of the process of forming a new thermodynamic phase. In contrast, new phases at continuous phase transitions start to form immedi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
SDS-PAGE
SDS-PAGE (sodium dodecyl sulfate–polyacrylamide gel electrophoresis) is a discontinuous electrophoretic system developed by Ulrich K. Laemmli which is commonly used as a method to separate proteins with molecular masses between 5 and 250 kDa. The combined use of sodium dodecyl sulfate (SDS, also known as sodium lauryl sulfate) and polyacrylamide gel allows to eliminate the influence of structure and charge, and proteins are separated solely on the basis of differences in their molecular weight. Properties SDS-PAGE is an electrophoresis method that allows protein separation by mass. The medium (also referred to as ′matrix′) is a polyacrylamide-based discontinuous gel. The polyacrylamide-gel is typically sandwiched between two glass plates in a slab gel. Although tube gels (in glass cylinders) were used historically, they were rapidly made obsolete with the invention of the more convenient slab gels. In addition, SDS (sodium dodecyl sulfate) is used. About 1.4 grams of S ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Fusion Protein
Fusion proteins or chimeric (kī-ˈmir-ik) proteins (literally, made of parts from different sources) are proteins created through the joining of two or more genes that originally coded for separate proteins. Translation of this ''fusion gene'' results in a single or multiple polypeptides with functional properties derived from each of the original proteins. ''Recombinant fusion proteins'' are created artificially by recombinant DNA technology for use in biological research or therapeutics. '' Chimeric'' or ''chimera'' usually designate hybrid proteins made of polypeptides having different functions or physico-chemical patterns. ''Chimeric mutant proteins'' occur naturally when a complex mutation, such as a chromosomal translocation, tandem duplication, or retrotransposition creates a novel coding sequence containing parts of the coding sequences from two different genes. Naturally occurring fusion proteins are commonly found in cancer cells, where they may function as oncoproteins ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Globular Protein
In biochemistry, globular proteins or spheroproteins are spherical ("globe-like") proteins and are one of the common protein types (the others being fibrous, disordered and membrane proteins). Globular proteins are somewhat water-soluble (forming colloids in water), unlike the fibrous or membrane proteins. There are multiple fold classes of globular proteins, since there are many different architectures that can fold into a roughly spherical shape. The term globin can refer more specifically to proteins including the globin fold. Globular structure and solubility The term globular protein is quite old (dating probably from the 19th century) and is now somewhat archaic given the hundreds of thousands of proteins and more elegant and descriptive structural motif vocabulary. The globular nature of these proteins can be determined without the means of modern techniques, but only by using ultracentrifuges or dynamic light scattering techniques. The spherical structure is induc ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Beta Sheet
The beta sheet, (β-sheet) (also β-pleated sheet) is a common motif of the regular protein secondary structure. Beta sheets consist of beta strands (β-strands) connected laterally by at least two or three backbone hydrogen bonds, forming a generally twisted, pleated sheet. A β-strand is a stretch of polypeptide chain typically 3 to 10 amino acids long with backbone in an extended conformation. The supramolecular association of β-sheets has been implicated in the formation of the fibrils and protein aggregates observed in amyloidosis, notably Alzheimer's disease. History The first β-sheet structure was proposed by William Astbury in the 1930s. He proposed the idea of hydrogen bonding between the peptide bonds of parallel or antiparallel extended β-strands. However, Astbury did not have the necessary data on the bond geometry of the amino acids in order to build accurate models, especially since he did not then know that the peptide bond was planar. A refined versi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Alpha Helix
The alpha helix (α-helix) is a common motif in the secondary structure of proteins and is a right hand-helix conformation in which every backbone N−H group hydrogen bonds to the backbone C=O group of the amino acid located four residues earlier along the protein sequence. The alpha helix is also called a classic Pauling–Corey–Branson α-helix. The name 3.613-helix is also used for this type of helix, denoting the average number of residues per helical turn, with 13 atoms being involved in the ring formed by the hydrogen bond. Among types of local structure in proteins, the α-helix is the most extreme and the most predictable from sequence, as well as the most prevalent. Discovery In the early 1930s, William Astbury showed that there were drastic changes in the X-ray fiber diffraction of moist wool or hair fibers upon significant stretching. The data suggested that the unstretched fibers had a coiled molecular structure with a characteristic repeat of ≈. Astb ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
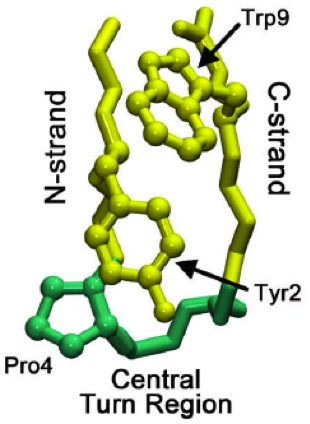
Beta Hairpin
The beta hairpin (sometimes also called beta-ribbon or beta-beta unit) is a simple protein structural motif involving two beta strands that look like a hairpin. The motif consists of two strands that are adjacent in primary structure, oriented in an antiparallel direction (the N-terminus of one sheet is adjacent to the C-terminus of the next), and linked by a short loop of two to five amino acids. Beta hairpins can occur in isolation or as part of a series of hydrogen bonded strands that collectively comprise a beta sheet. Researchers such as Francisco Blanco ''et al.'' have used protein NMR to show that beta-hairpins can be formed from isolated short peptides in aqueous solution, suggesting that hairpins could form nucleation sites for protein folding. Classification Beta hairpins were originally categorized solely by the number of amino acid residues in their loop sequences, such that they were named one-residue, two-residue, etc. This system, however, is somewhat ambiguous ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Amino Acid
Amino acids are organic compounds that contain both amino and carboxylic acid functional groups. Although hundreds of amino acids exist in nature, by far the most important are the alpha-amino acids, which comprise proteins. Only 22 alpha amino acids appear in the genetic code. Amino acids can be classified according to the locations of the core structural functional groups, as Alpha and beta carbon, alpha- , beta- , gamma- or delta- amino acids; other categories relate to Chemical polarity, polarity, ionization, and side chain group type (aliphatic, Open-chain compound, acyclic, aromatic, containing hydroxyl or sulfur, etc.). In the form of proteins, amino acid '' residues'' form the second-largest component (water being the largest) of human muscles and other tissues. Beyond their role as residues in proteins, amino acids participate in a number of processes such as neurotransmitter transport and biosynthesis. It is thought that they played a key role in enabling life ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Hydrophobe
In chemistry, hydrophobicity is the physical property of a molecule that is seemingly repelled from a mass of water (known as a hydrophobe). In contrast, hydrophiles are attracted to water. Hydrophobic molecules tend to be nonpolar and, thus, prefer other neutral molecules and nonpolar solvents. Because water molecules are polar, hydrophobes do not dissolve well among them. Hydrophobic molecules in water often cluster together, forming micelles. Water on hydrophobic surfaces will exhibit a high contact angle. Examples of hydrophobic molecules include the alkanes, oils, fats, and greasy substances in general. Hydrophobic materials are used for oil removal from water, the management of oil spills, and chemical separation processes to remove non-polar substances from polar compounds. Hydrophobic is often used interchangeably with lipophilic, "fat-loving". However, the two terms are not synonymous. While hydrophobic substances are usually lipophilic, there are exceptions, suc ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ab Initio Quantum Chemistry Methods
''Ab initio'' quantum chemistry methods are computational chemistry methods based on quantum chemistry. The term was first used in quantum chemistry by Robert Parr and coworkers, including David Craig in a semiempirical study on the excited states of benzene. The background is described by Parr. ''Ab initio'' means "from first principles" or "from the beginning", implying that the only inputs into an ''ab initio'' calculation are physical constants. ''Ab initio'' quantum chemistry methods attempt to solve the electronic Schrödinger equation given the positions of the nuclei and the number of electrons in order to yield useful information such as electron densities, energies and other properties of the system. The ability to run these calculations has enabled theoretical chemists to solve a range of problems and their importance is highlighted by the awarding of the Nobel prize to John Pople and Walter Kohn. Accuracy and scaling ''Ab initio'' electronic structure method ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein
Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residues. Proteins perform a vast array of functions within organisms, including catalysing metabolic reactions, DNA replication, responding to stimuli, providing structure to cells and organisms, and transporting molecules from one location to another. Proteins differ from one another primarily in their sequence of amino acids, which is dictated by the nucleotide sequence of their genes, and which usually results in protein folding into a specific 3D structure that determines its activity. A linear chain of amino acid residues is called a polypeptide. A protein contains at least one long polypeptide. Short polypeptides, containing less than 20–30 residues, are rarely considered to be proteins and are commonly called peptides. The individual amino acid residues are bonded together by peptide bonds and adjacent amino acid residues. The sequence of amino acid residue ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |