|
Elongation Factor G
EF-G (elongation factor G, historically known as translocase) is a prokaryotic elongation factor involved in protein translation. As a GTPase, EF-G catalyzes the movement (translocation) of transfer RNA (tRNA) and messenger RNA (mRNA) through the ribosome. Structure Encoded by the ''fusA'' gene on the ''str'' operon, EF-G is made up of 704 amino acids that form 5 domains, labeled Domain I through Domain V. Domain I may be referred to as the G-domain or as Domain I(G), since it binds to and hydrolyzes guanosine triphosphate (GTP). Domain I also helps EF-G bind to the ribosome, and contains the N-terminal of the polypeptide chain. Domain IV is important for translocation, as it undergoes a significant conformational change and enters the A site on the 30S ribosomal subunit, pushing the mRNA and tRNA molecules from the A site to the P site. The five domains may be also separated into two super-domains. Super-domain I consists of Domains I and II, and super-domain II consists o ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Prokaryotic Elongation Factor
Elongation factors are a set of proteins that function at the ribosome, during protein synthesis, to facilitate translational elongation from the formation of the first to the last peptide bond of a growing polypeptide. Most common elongation factors in prokaryotes are EF-Tu, EF-Ts, EF-G. Bacteria and eukaryotes use elongation factors that are largely homologous to each other, but with distinct structures and different research nomenclatures. Elongation is the most rapid step in translation. In bacteria, it proceeds at a rate of 15 to 20 amino acids added per second (about 45-60 nucleotides per second). In eukaryotes the rate is about two amino acids per second (about 6 nucleotides read per second). Elongation factors play a role in orchestrating the events of this process, and in ensuring the high accuracy translation at these speeds. Nomenclature of homologous EFs In addition to their cytoplasmic machinery, eukaryotic mitochondria and plastids have their own translat ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Peptidyl Transferase
The peptidyl transferase is an aminoacyltransferase () as well as the primary enzymatic function of the ribosome, which forms peptide bonds between adjacent amino acids using tRNAs during the translation process of protein biosynthesis. The substrates for the peptidyl transferase reaction are two tRNA molecules, one bearing the growing peptide chain and the other bearing the amino acid that will be added to the chain. The peptidyl chain and the amino acids are attached to their respective tRNAs via ester bonds to the O atom at the CCA-3' ends of these tRNAs. Peptidyl transferase is an enzyme that catalyzes the addition of an amino acid residue in order to grow the polypeptide chain in protein synthesis. It is located in the large ribosomal subunit, where it catalyzes the peptide bond formation. It is composed entirely of RNA. The alignment between the CCA ends of the ribosome-bound peptidyl tRNA and aminoacyl tRNA in the peptidyl transferase center contribute to its ability to cat ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Domain (biology)
In biological taxonomy, a domain ( or ) (Latin: ''regio''), also dominion, superkingdom, realm, or empire, is the highest taxonomic rank of all organisms taken together. It was introduced in the three-domain system of taxonomy devised by Carl Woese, Otto Kandler and Mark Wheelis in 1990. According to the domain system, the tree of life consists of either three domains such as Archaea, Bacteria, and Eukarya, or two domains consisting of Archaea and Bacteria, with Eukarya included in Archaea. The first two are all prokaryotes, single-celled microorganisms without a membrane-bound nucleus. All organisms that have a cell nucleus and other membrane-bound organelles are included in Eukarya. Non-cellular life is not included in this system. Alternatives to the three-domain system include the earlier two-empire system (with the empires Prokaryota and Eukaryota), and the eocyte hypothesis (with two domains of Bacteria and Archaea, with Eukarya included as a branch of Archaea). Term ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
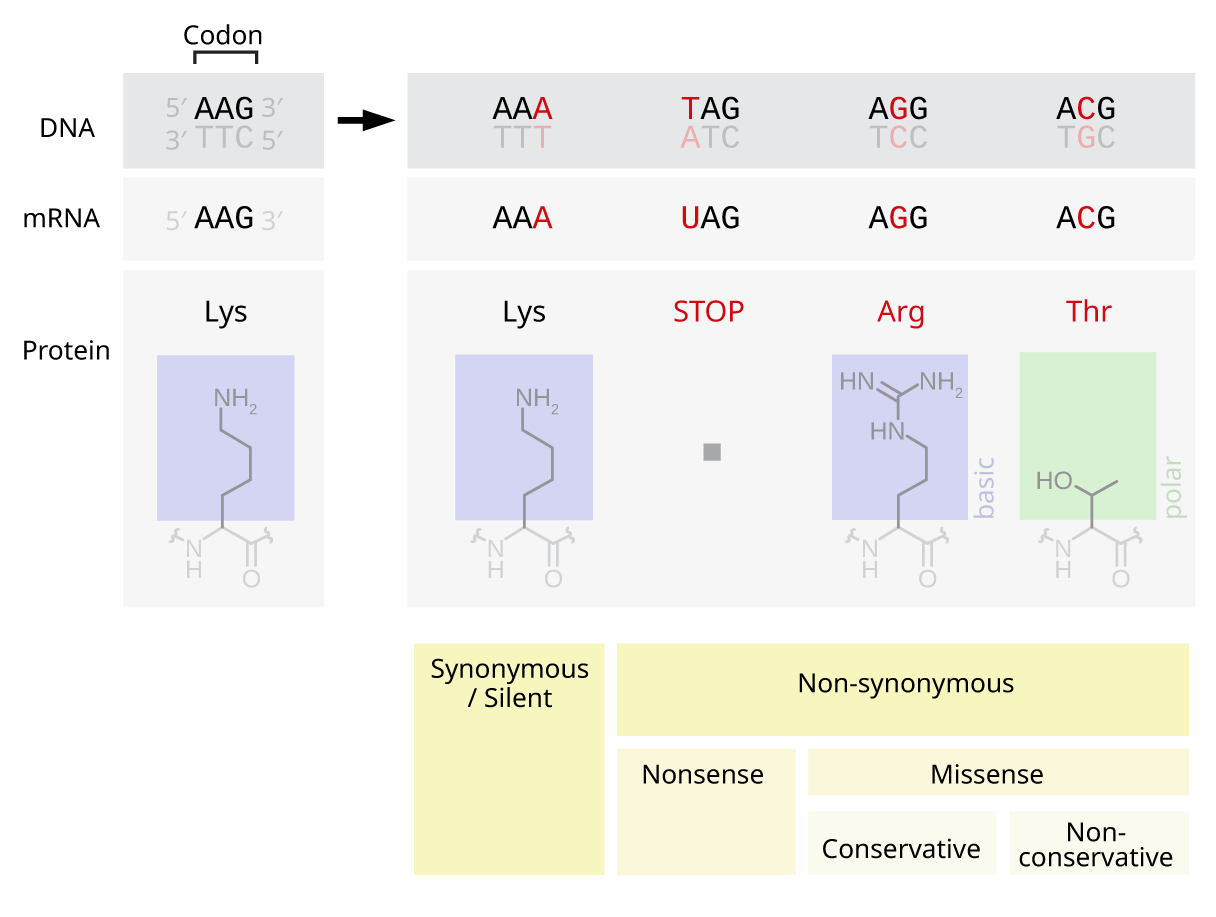
Point Mutation
A point mutation is a genetic mutation where a single nucleotide base is changed, inserted or deleted from a DNA or RNA sequence of an organism's genome. Point mutations have a variety of effects on the downstream protein product—consequences that are moderately predictable based upon the specifics of the mutation. These consequences can range from no effect (e.g. synonymous mutations) to deleterious effects (e.g. frameshift mutations), with regard to protein production, composition, and function. Causes Point mutations usually take place during DNA replication. DNA replication occurs when one double-stranded DNA molecule creates two single strands of DNA, each of which is a template for the creation of the complementary strand. A single point mutation can change the whole DNA sequence. Changing one purine or pyrimidine may change the amino acid that the nucleotides code for. Point mutations may arise from spontaneous mutations that occur during DNA replication. The rate of ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Bacteria
Bacteria (; singular: bacterium) are ubiquitous, mostly free-living organisms often consisting of one biological cell. They constitute a large domain of prokaryotic microorganisms. Typically a few micrometres in length, bacteria were among the first life forms to appear on Earth, and are present in most of its habitats. Bacteria inhabit soil, water, acidic hot springs, radioactive waste, and the deep biosphere of Earth's crust. Bacteria are vital in many stages of the nutrient cycle by recycling nutrients such as the fixation of nitrogen from the atmosphere. The nutrient cycle includes the decomposition of dead bodies; bacteria are responsible for the putrefaction stage in this process. In the biological communities surrounding hydrothermal vents and cold seeps, extremophile bacteria provide the nutrients needed to sustain life by converting dissolved compounds, such as hydrogen sulphide and methane, to energy. Bacteria also live in symbiotic and parasitic relationsh ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Staphylococcus Aureus
''Staphylococcus aureus'' is a Gram-positive spherically shaped bacterium, a member of the Bacillota, and is a usual member of the microbiota of the body, frequently found in the upper respiratory tract and on the skin. It is often positive for catalase and nitrate reduction and is a facultative anaerobe that can grow without the need for oxygen. Although ''S. aureus'' usually acts as a commensal of the human microbiota, it can also become an opportunistic pathogen, being a common cause of skin infections including abscesses, respiratory infections such as sinusitis, and food poisoning. Pathogenic strains often promote infections by producing virulence factors such as potent protein toxins, and the expression of a cell-surface protein that binds and inactivates antibodies. ''S. aureus'' is one of the leading pathogens for deaths associated with antimicrobial resistance and the emergence of antibiotic-resistant strains, such as methicillin-resistant ''S. aureus'' (MRSA ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
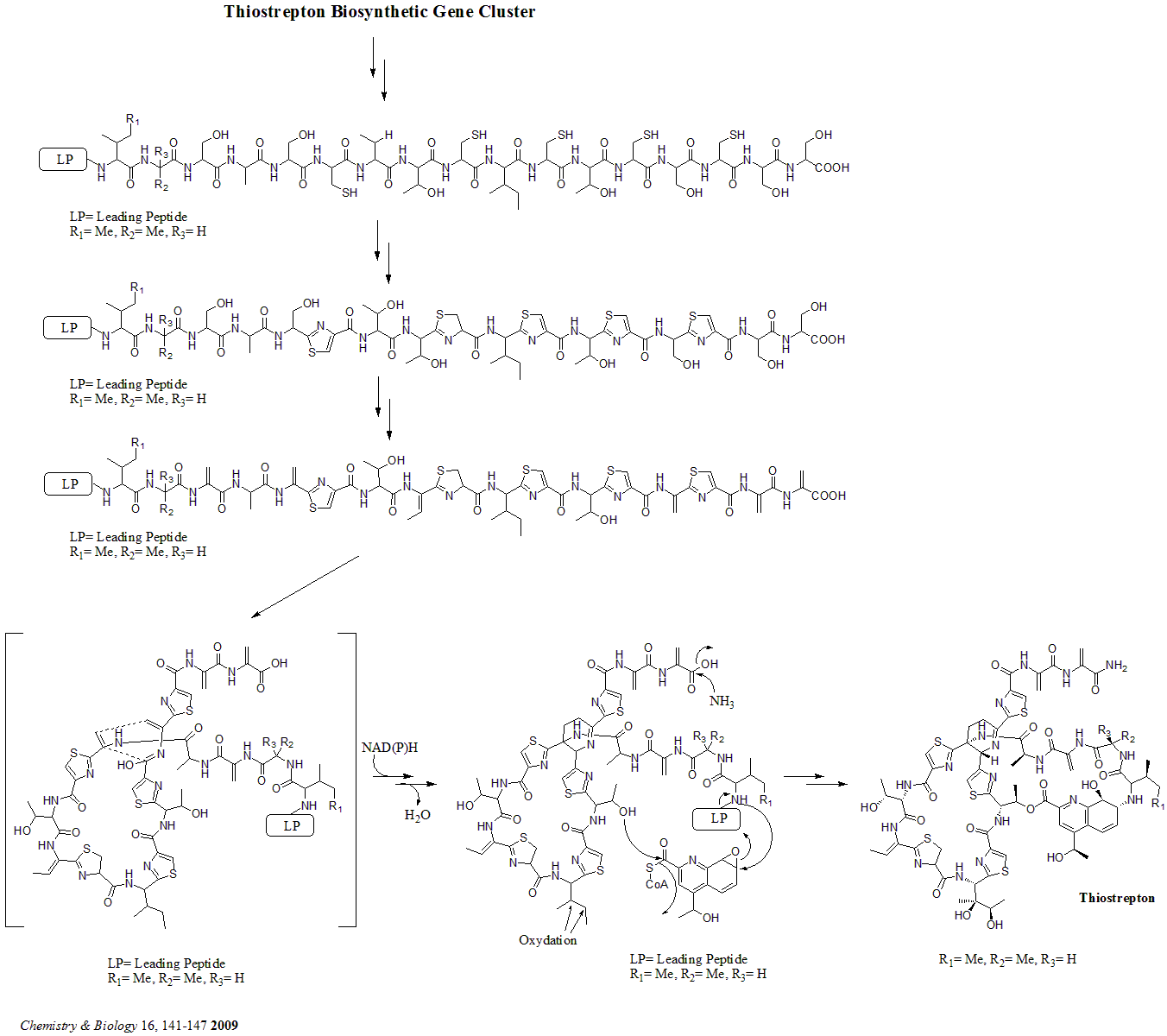
Thiostrepton
Thiostrepton is a natural cyclic oligopeptide antibiotic of the thiopeptide class, derived from several strains of streptomycetes, such as '' Streptomyces azureus'' and '' Streptomyces laurentii''. Thiostrepton is a natural product of the ribosomally synthesized and post-translationally modified peptide (RiPP) class. History Thiostrepton was discovered by Donovick ''et al.'' who described its antibacterial properties in 1955. Dorothy Crowfoot Hodgkin solved the structure of thiostrepton in 1970. Early in 1978, Bycroft and Gowland proposed the biosynthesis of thiostrepton, which was still unclear until 2009. Several studies of thiopeptide biosynthesis have been contemporarily published in 2009 and two of them (Liao ''et al.'' and Kelly ''et al.'') included the similar biosynthesis of thiostrepton: it's ribosomally synthesized from thiostrepton biosynthetic genes (tsr genes) and posttranslational modification is needed. A total synthesis of thiostrepton was completed by K.C. ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Antibiotics
An antibiotic is a type of antimicrobial substance active against bacteria. It is the most important type of antibacterial agent for fighting bacterial infections, and antibiotic medications are widely used in the treatment and prevention of such infections. They may either kill or inhibit the growth of bacteria. A limited number of antibiotics also possess antiprotozoal activity. Antibiotics are not effective against viruses such as the common cold or influenza; drugs which inhibit viruses are termed antiviral drugs or antivirals rather than antibiotics. Sometimes, the term ''antibiotic''—literally "opposing life", from the Greek roots ἀντι ''anti'', "against" and βίος ''bios'', "life"—is broadly used to refer to any substance used against microbes, but in the usual medical usage, antibiotics (such as penicillin) are those produced naturally (by one microorganism fighting another), whereas non-antibiotic antibacterials (such as sulfonamides and antisep ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Pathogenic Bacteria
Pathogenic bacteria are bacteria that can cause disease. This article focuses on the bacteria that are pathogenic to humans. Most species of bacteria are harmless and are often Probiotic, beneficial but others can cause infectious diseases. The number of these pathogenic species in humans is estimated to be fewer than a hundred. By contrast, several thousand species are part of the gut flora present in the digestive tract. The body is continually exposed to many species of bacteria, including beneficial commensals, which grow on the skin and mucous membranes, and saprophytes, which grow mainly in the soil and in decomposition, decaying matter. The blood and tissue fluids contain nutrients sufficient to sustain the growth of many bacteria. The body has defence mechanisms that enable it to resist microbial invasion of its tissues and give it a natural immune system, immunity or innate immunity, innate resistance against many microorganisms. Pathogenic bacteria are specially adapt ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Prokaryotic Initiation Factor-3
In molecular biology, translation initiation factor IF-3 (gene infC) is one of the three factors required for the initiation of protein biosynthesis in bacteria. IF-3 is thought to function as a fidelity factor during the assembly of the ternary initiation complex which consists of the 30S ribosomal subunit, the initiator tRNA and the messenger RNA. IF-3 is a basic protein that binds to the 30S ribosomal subunit. The chloroplast homolog enhances the poly( A, U, G)-dependent binding of the initiator tRNA to its ribosomal 30s subunits. IF1–IF3 may also perform ribosome recycling. IF3 is not universally found in all bacterial species. However, in '' E. coli'', it is required for the 30S subunit to bind to the initiation site in mRNA. In addition, it has several other jobs including the stabilization of free 30S subunits, enables 30S subunits to bind to mRNA and checks for accuracy against the first aminoacyl-tRNA. It also allows for rapid codon-anticodon pairing for the initiato ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ribosome Recycling Factor
Ribosome recycling factor or ribosome release factor (RRF) is a protein found in bacterial cells as well as eukaryotic organelles, specifically mitochondria and chloroplasts. It functions to recycle ribosomes after completion of protein synthesis (bacterial translation). In humans, the mitochrondrial version is coded by the MRRF gene. Discovery The ribosome recycling factor was discovered in the early 1970s by the work of Akira Kaji and Akikazu Hiroshima at the University of Pennsylvania. Their work described the requirement for two protein factors to release ribosomes from mRNA. These two factors were identified as RRF, an unknown protein until then, and Elongation Factor G (EF-G), a protein already identified and known to function in protein synthesis. RRF was originally called Ribosome ''Releasing'' Factor but is now called Ribosome ''Recycling'' Factor. Function RRF accomplishes the recycling of ribosomes by splitting ribosomes into subunits, thereby releasing the bound m ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
.jpg)