|
EQTL
Expression quantitative trait loci (eQTLs) are genomic loci that explain variation in expression levels of mRNAs. Distant and local, trans- and cis-eQTLs, respectively An expression quantitative trait is an amount of an mRNA transcript or a protein. These are usually the product of a single gene with a specific chromosomal location. This distinguishes expression quantitative traits from most complex traits, which are not the product of the expression of a single gene. Chromosomal loci that explain variance in expression traits are called eQTLs. eQTLs located near the gene-of-origin (gene which produces the transcript or protein) are referred to as local eQTLs or cis-eQTLs. By contrast, those located distant from their gene of origin, often on different chromosomes, are referred to as distant eQTLs or trans-eQTLs. The first genome-wide study of gene expression was carried out in yeast and published in 2002. The initial wave of eQTL studies employed microarrays to measure genome ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
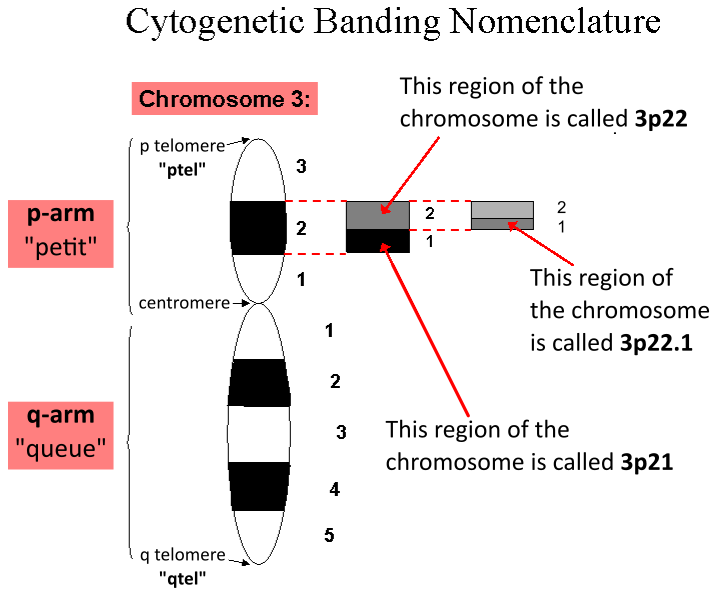
Locus (genetics)
In genetics, a locus (plural loci) is a specific, fixed position on a chromosome where a particular gene or genetic marker is located. Each chromosome carries many genes, with each gene occupying a different position or locus; in humans, the total number of protein-coding genes in a complete haploid set of 23 chromosomes is estimated at 19,000–20,000. Genes may possess multiple variants known as alleles, and an allele may also be said to reside at a particular locus. Diploid and polyploid cells whose chromosomes have the same allele at a given locus are called homozygous with respect to that locus, while those that have different alleles at a given locus are called heterozygous. The ordered list of loci known for a particular genome is called a gene map. Gene mapping is the process of determining the specific locus or loci responsible for producing a particular phenotype or biological trait. Association mapping, also known as "linkage disequilibrium mapping", is a method of ma ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Genetic Variation
Genetic variation is the difference in DNA among individuals or the differences between populations. The multiple sources of genetic variation include mutation and genetic recombination. Mutations are the ultimate sources of genetic variation, but other mechanisms, such as genetic drift, contribute to it, as well. Among individuals within a population Genetic variation can be identified at many levels. Identifying genetic variation is possible from observations of phenotypic variation in either quantitative traits (traits that vary continuously and are coded for by many genes (e.g., leg length in dogs)) or discrete traits (traits that fall into discrete categories and are coded for by one or a few genes (e.g., white, pink, or red petal color in certain flowers)). Genetic variation can also be identified by examining variation at the level of enzymes using the process of protein electrophoresis. Polymorphic genes have more than one allele at each locus. Half of the genes ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Classical Genetics
Classical genetics is the branch of genetics based solely on visible results of reproductive acts. It is the oldest discipline in the field of genetics, going back to the experiments on Mendelian inheritance by Gregor Mendel who made it possible to identify the basic mechanisms of heredity. Subsequently, these mechanisms have been studied and explained at the molecular level. Classical genetics consists of the techniques and methodologies of genetics that were in use before the advent of molecular biology. A key discovery of classical genetics in eukaryotes was genetic linkage. The observation that some genes do not segregate independently at meiosis broke the laws of Mendelian inheritance and provided science with a way to map characteristics to a location on the chromosomes. Linkage maps are still used today, especially in breeding for plant improvement. After the discovery of the genetic code and such tools of cloning as restriction enzymes, the avenues of investigation open ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
TWAS V1
Transcriptome-wide association study (TWAS) is a statistical genetics methodology to improve detection power and provide functional annotation for genetic associations with phenotypes by integrating single-nucleotide polymorphism to trait (SNP-trait) associations from genome-wide association studies with SNP-based prediction models of gene expression. The approach was presented by Eric R. Gamazon et al. and Alexander Gusev et al. in the journal ''Nature Genetics''. This methodology has been widely adopted, having received 2057 citations (as of December 24, 2021) according to Google Scholar. See also * Genome-wide association study * Functional genomics * Genotype-Tissue Expression (GTEx) project * Epigenome-wide association study An epigenome-wide association study (EWAS) is an examination of a genome-wide set of quantifiable epigenetic marks, such as DNA methylation, in different individuals to derive associations between epigenetic variation and a particular identifiab ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Quantitative Trait Locus
A quantitative trait locus (QTL) is a locus (section of DNA) that correlates with variation of a quantitative trait in the phenotype of a population of organisms. QTLs are mapped by identifying which molecular markers (such as SNPs or AFLPs) correlate with an observed trait. This is often an early step in identifying the actual genes that cause the trait variation. Definition A quantitative trait locus (QTL) is a region of DNA which is associated with a particular phenotypic trait, which varies in degree and which can be attributed to polygenic effects, i.e., the product of two or more genes, and their environment. . These QTLs are often found on different chromosomes. The number of QTLs which explain variation in the phenotypic trait indicates the genetic architecture of a trait. It may indicate that plant height is controlled by many genes of small effect, or by a few genes of large effect. Typically, QTLs underlie continuous traits (those traits which vary continuously, ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Epigenome-wide Association Study
An epigenome-wide association study (EWAS) is an examination of a genome-wide set of quantifiable epigenetic marks, such as DNA methylation, in different individuals to derive associations between epigenetic variation and a particular identifiable phenotype/trait. When patterns change such as DNA methylation at specific loci, discriminating the phenotypically affected cases from control individuals, this is considered an indication that epigenetic perturbation has taken place that is associated, causally or consequentially, with the phenotype. Background The epigenome is governed by both genetic and environmental factors, causing it to be highly dynamic and complex. Epigenetic information exists in the cell as DNA and histone marks, as well as non-coding RNAs. DNA methylation (DNAm) patterns change over time, and vary between developmental stage and tissue type. The main type of DNAm is at cytosines within CpG dinucleotides which is known to be involved in gene expression reg ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
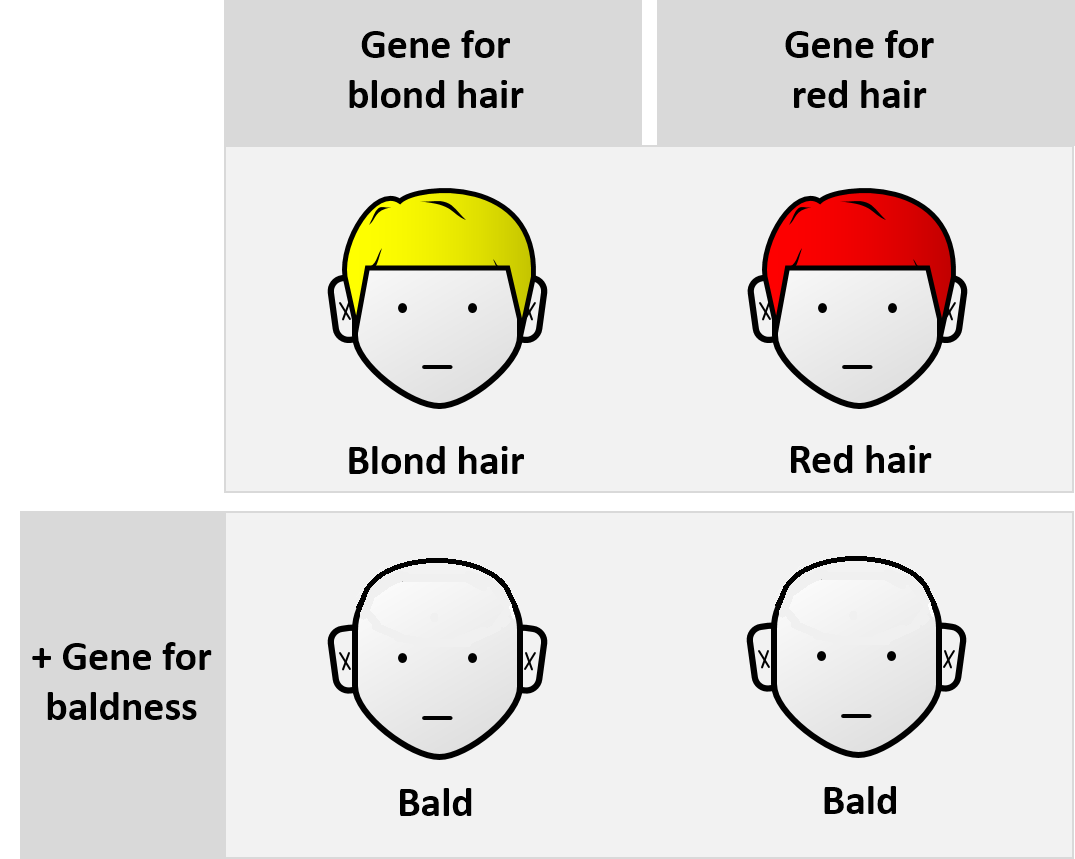
Epistasis
Epistasis is a phenomenon in genetics in which the effect of a gene mutation is dependent on the presence or absence of mutations in one or more other genes, respectively termed modifier genes. In other words, the effect of the mutation is dependent on the genetic background in which it appears. Epistatic mutations therefore have different effects on their own than when they occur together. Originally, the term ''epistasis'' specifically meant that the effect of a gene variant is masked by that of a different gene. The concept of ''epistasis'' originated in genetics in 1907 but is now used in biochemistry, computational biology and evolutionary biology. The phenomenon arises due to interactions, either between genes (such as mutations also being needed in regulators of gene expression) or within them (multiple mutations being needed before the gene loses function), leading to non-linear effects. Epistasis has a great influence on the shape of evolutionary landscapes, which l ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
GeneNetwork
GeneNetwork is a combined database and open-source software, open-source bioinformatics data analysis software resource for systems genetics. This resource is used to study gene regulatory networks that link DNA sequence differences to corresponding differences in gene and protein expression and to variation in traits such as health and disease risk. Data sets in GeneNetwork are typically made up of large collections of genotypes (e.g., single-nucleotide polymorphism, SNPs) and phenotypes from groups of individuals, including humans, strains of mice and rats, and organisms as diverse as Drosophila melanogaster, Arabidopsis thaliana, and barley. The inclusion of genotypes makes it practical to carry out web-based gene mapping to discover those regions of genomes that contribute to differences among individuals in mRNA, protein, and metabolite levels, as well as differences in cell function, anatomy, physiology, and behavior. History Development of GeneNetwork started at the Unive ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Alexander Gusev (scientist)
Alexander (Sasha) Gusev is a computational biologist and an Assistant Professor of Medicine at Harvard Medical School. Research and career Alexander Gusev has developed computational methods that use genetic data to decipher disease mechanisms. For example, he has identified 34 new genes associated with increased risk of earliest-stage ovarian cancer. He has developed computational methods that integrate molecular data to facilitate functional interpretation of findings from genome-wide association studies. He has contributed to the development of the transcriptome-wide association study approach to mapping disease-associated genes. In addition, he studies the interactions between germline In biology and genetics, the germline is the population of a multicellular organism's cells that pass on their genetic material to the progeny (offspring). In other words, they are the cells that form the egg, sperm and the fertilised egg. They ... (host) and somatic events (tumor) - which ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Transcriptome-wide Association Study
Transcriptome-wide association study (TWAS) is a statistical genetics methodology to improve detection power and provide functional annotation for genetic associations with phenotypes by integrating single-nucleotide polymorphism to trait (SNP-trait) associations from genome-wide association studies with SNP-based prediction models of gene expression. The approach was presented by Eric R. Gamazon et al. and Alexander Gusev et al. in the journal ''Nature Genetics''. This methodology has been widely adopted, having received 2057 citations (as of December 24, 2021) according to Google Scholar. See also * Genome-wide association study * Functional genomics * Genotype-Tissue Expression (GTEx) project * Epigenome-wide association study An epigenome-wide association study (EWAS) is an examination of a genome-wide set of quantifiable epigenetic marks, such as DNA methylation, in different individuals to derive associations between epigenetic variation and a particular identifiable ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Eric R
The given name Eric, Erich, Erikk, Erik, Erick, or Eirik is derived from the Old Norse name ''Eiríkr'' (or ''Eríkr'' in Old East Norse due to monophthongization). The first element, ''ei-'' may be derived from the older Proto-Norse ''* aina(z)'', meaning "one, alone, unique", ''as in the form'' ''Æ∆inrikr'' explicitly, but it could also be from ''* aiwa(z)'' "everlasting, eternity", as in the Gothic form ''Euric''. The second element ''- ríkr'' stems either from Proto-Germanic ''* ríks'' "king, ruler" (cf. Gothic ''reiks'') or the therefrom derived ''* ríkijaz'' "kingly, powerful, rich, prince"; from the common Proto-Indo-European root * h₃rḗǵs. The name is thus usually taken to mean "sole ruler, autocrat" or "eternal ruler, ever powerful". ''Eric'' used in the sense of a proper noun meaning "one ruler" may be the origin of ''Eriksgata'', and if so it would have meant "one ruler's journey". The tour was the medieval Swedish king's journey, when newly elected, to s ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Single Nucleotide Polymorphism
In genetics, a single-nucleotide polymorphism (SNP ; plural SNPs ) is a germline substitution of a single nucleotide at a specific position in the genome. Although certain definitions require the substitution to be present in a sufficiently large fraction of the population (e.g. 1% or more), many publications do not apply such a frequency threshold. For example, at a specific base position in the human genome, the G nucleotide may appear in most individuals, but in a minority of individuals, the position is occupied by an A. This means that there is a SNP at this specific position, and the two possible nucleotide variations – G or A – are said to be the alleles for this specific position. SNPs pinpoint differences in our susceptibility to a wide range of diseases, for example age-related macular degeneration (a common SNP in the CFH gene is associated with increased risk of the disease) or nonalcoholic fatty liver disease (a SNP in the PNPLA3 gene is associated with incr ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |