|
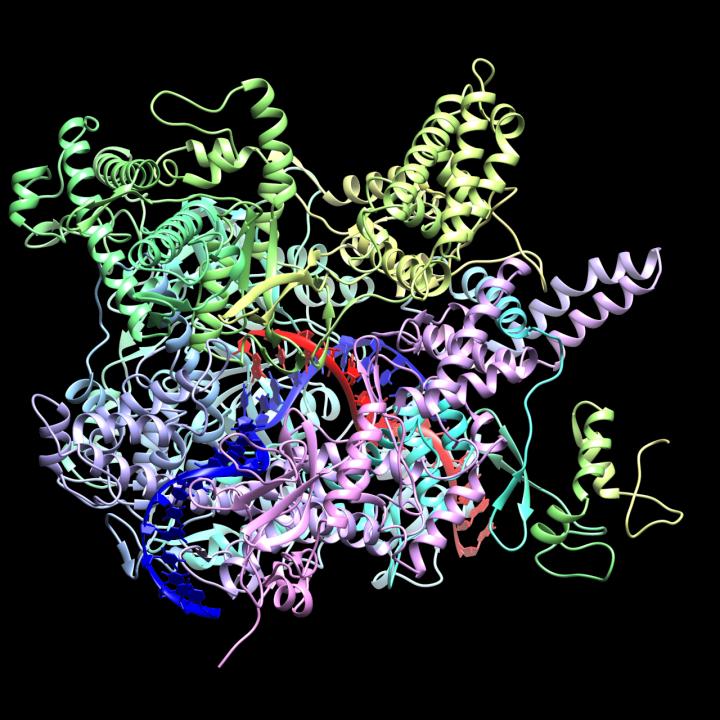
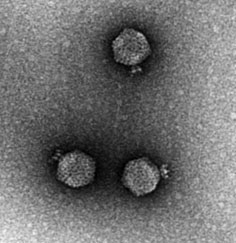
CrAssphage
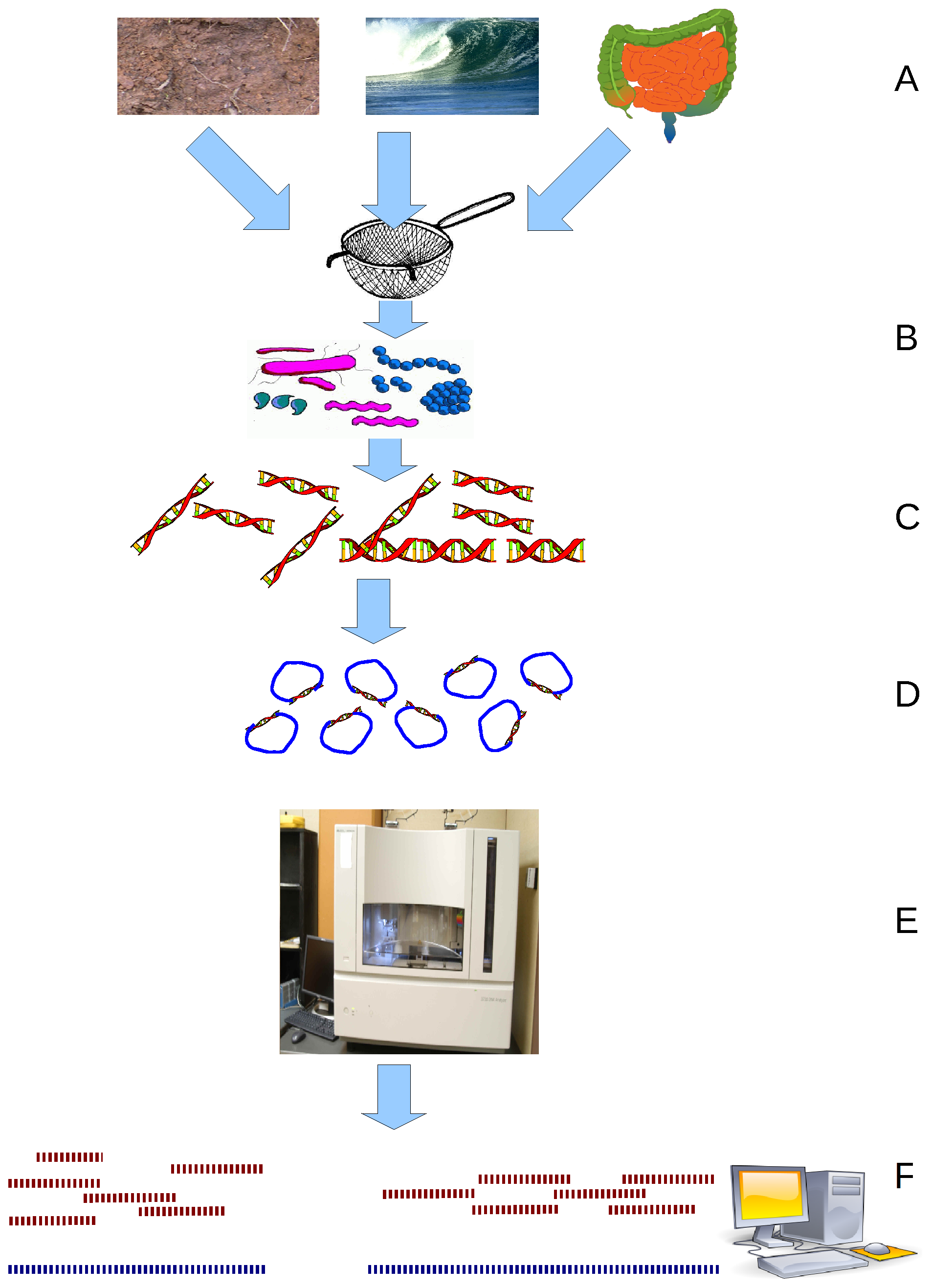
CrAss-like phage are a bacteriophage (virus that infects bacteria) family that was discovered in 2014 by cross assembling reads in human fecal metagenomes. In silico comparative genomics and taxonomic analysis have found that crAss-like phages represent a highly abundant and diverse family of viruses. CrAss-like phage were predicted to infect bacteria of the ''Bacteroidota'' phylum and the prediction was later confirmed when the first crAss-like phage (crAss001) was isolated on a ''Bacteroidota'' host (''B. intestinalis'') in 2018. The presence of crAss-like phage in the human gut microbiota is not yet associated with any health condition. Discovery The crAss (cross-assembly) software used to discover the first crAss-like phage, p-crAssphage (prototypical-crAssphage), relies on cross assembling reads from multiple metagenomes obtained from the same environment. The goal of cross-assembly is that unknown reads from one metagenome align with known reads, or reads that have similarity ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Bacteriophages
A bacteriophage (), also known informally as a ''phage'' (), is a duplodnaviria virus that infects and replicates within bacteria and archaea. The term was derived from "bacteria" and the Greek φαγεῖν ('), meaning "to devour". Bacteriophages are composed of proteins that encapsulate a DNA or RNA genome, and may have structures that are either simple or elaborate. Their genomes may encode as few as four genes (e.g. MS2) and as many as hundreds of genes. Phages replicate within the bacterium following the injection of their genome into its cytoplasm. Bacteriophages are among the most common and diverse entities in the biosphere. Bacteriophages are ubiquitous viruses, found wherever bacteria exist. It is estimated there are more than 1031 bacteriophages on the planet, more than every other organism on Earth, including bacteria, combined. Viruses are the most abundant biological entity in the water column of the world's oceans, and the second largest component of biomass ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Bacteriophage
A bacteriophage (), also known informally as a ''phage'' (), is a duplodnaviria virus that infects and replicates within bacteria and archaea. The term was derived from "bacteria" and the Greek φαγεῖν ('), meaning "to devour". Bacteriophages are composed of proteins that encapsulate a DNA or RNA genome, and may have structures that are either simple or elaborate. Their genomes may encode as few as four genes (e.g. MS2) and as many as hundreds of genes. Phages replicate within the bacterium following the injection of their genome into its cytoplasm. Bacteriophages are among the most common and diverse entities in the biosphere. Bacteriophages are ubiquitous viruses, found wherever bacteria exist. It is estimated there are more than 1031 bacteriophages on the planet, more than every other organism on Earth, including bacteria, combined. Viruses are the most abundant biological entity in the water column of the world's oceans, and the second largest component of biom ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Metagenome
Metagenomics is the study of genetic material recovered directly from environmental or clinical samples by a method called sequencing. The broad field may also be referred to as environmental genomics, ecogenomics, community genomics or microbiomics. While traditional microbiology and microbial genome sequencing and genomics rely upon cultivated clonal cultures, early environmental gene sequencing cloned specific genes (often the 16S rRNA gene) to produce a profile of diversity in a natural sample. Such work revealed that the vast majority of microbial biodiversity had been missed by cultivation-based methods. Because of its ability to reveal the previously hidden diversity of microscopic life, metagenomics offers a powerful lens for viewing the microbial world that has the potential to revolutionize understanding of the entire living world. As the price of DNA sequencing continues to fall, metagenomics now allows microbial ecology to be investigated at a much greater scale ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Comparative Genomics
Comparative genomics is a field of biological research in which the genomic features of different organisms are compared. The genomic features may include the DNA sequence, genes, gene order, regulatory sequences, and other genomic structural landmarks. In this branch of genomics, whole or large parts of genomes resulting from genome projects are compared to study basic biological similarities and differences as well as evolutionary relationships between organisms. The major principle of comparative genomics is that common features of two organisms will often be encoded within the DNA that is evolutionarily conserved between them. Therefore, comparative genomic approaches start with making some form of alignment of genome sequences and looking for orthologous sequences (sequences that share a common ancestry) in the aligned genomes and checking to what extent those sequences are conserved. Based on these, genome and molecular evolution are inferred and this may in turn be put in ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Open Reading Frame
In molecular biology, open reading frames (ORFs) are defined as spans of DNA sequence between the start and stop codons. Usually, this is considered within a studied region of a prokaryotic DNA sequence, where only one of the six possible reading frames will be "open" (the "reading", however, refers to the RNA produced by transcription of the DNA and its subsequent interaction with the ribosome in translation). Such an ORF may contain a start codon (usually AUG in terms of RNA) and by definition cannot extend beyond a stop codon (usually UAA, UAG or UGA in RNA). That start codon (not necessarily the first) indicates where translation may start. The transcription termination site is located after the ORF, beyond the translation stop codon. If transcription were to cease before the stop codon, an incomplete protein would be made during translation. In eukaryotic genes with multiple exons, introns are removed and exons are then joined together after transcription to yield the final ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Bacteroidota
The phylum Bacteroidota (synonym Bacteroidetes) is composed of three large classes of Gram-negative, nonsporeforming, anaerobic or aerobic, and rod-shaped bacteria that are widely distributed in the environment, including in soil, sediments, and sea water, as well as in the guts and on the skin of animals. Although some ''Bacteroides'' spp. can be opportunistic pathogens, many ''Bacteroidota'' are symbiotic species highly adjusted to the gastrointestinal tract. ''Bacteroides'' are highly abundant in intestines, reaching up to 1011 cells g−1 of intestinal material. They perform metabolic conversions that are essential for the host, such as degradation of proteins or complex sugar polymers. ''Bacteroidota'' colonize the gastrointestinal tract already in infants, as non-digestible oligosaccharides in mother milk support the growth of both ''Bacteroides'' and ''Bifidobacterium'' spp. ''Bacteroides'' spp. are selectively recognized by the immune system of the host through specific ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Indicator Bacteria
Indicator bacteria are types of bacteria used to detect and estimate the level of fecal contamination of water. They are not dangerous to human health but are used to indicate the presence of a health risk. Each gram of human feces contains approximately ~100 billion () bacteria. These bacteria may include species of pathogenic bacteria, such as '' Salmonella'' or '' Campylobacter'', associated with gastroenteritis. In addition, feces may contain pathogenic viruses, protozoa and parasites. Fecal material can enter the environment from many sources including waste water treatment plants, livestock or poultry manure, sanitary landfills, septic systems, sewage sludge, pets and wildlife. If sufficient quantities are ingested, fecal pathogens can cause disease. The variety and often low concentrations of pathogens in environmental waters makes them difficult to test for individually. Public agencies therefore use the presence of other more abundant and more easily detected fecal b ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
RNA Polymerase
In molecular biology, RNA polymerase (abbreviated RNAP or RNApol), or more specifically DNA-directed/dependent RNA polymerase (DdRP), is an enzyme that synthesizes RNA from a DNA template. Using the enzyme helicase, RNAP locally opens the double-stranded DNA so that one strand of the exposed nucleotides can be used as a template for the synthesis of RNA, a process called transcription. A transcription factor and its associated transcription mediator complex must be attached to a DNA binding site called a promoter region before RNAP can initiate the DNA unwinding at that position. RNAP not only initiates RNA transcription, it also guides the nucleotides into position, facilitates attachment and elongation, has intrinsic proofreading and replacement capabilities, and termination recognition capability. In eukaryotes, RNAP can build chains as long as 2.4 million nucleotides. RNAP produces RNA that, functionally, is either for protein coding, i.e. messenger RNA (mRNA); or n ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
RNA Interference
RNA interference (RNAi) is a biological process in which RNA molecules are involved in sequence-specific suppression of gene expression by double-stranded RNA, through translational or transcriptional repression. Historically, RNAi was known by other names, including ''co-suppression'', ''post-transcriptional gene silencing'' (PTGS), and ''quelling''. The detailed study of each of these seemingly different processes elucidated that the identity of these phenomena were all actually RNAi. Andrew Fire and Craig C. Mello shared the 2006 Nobel Prize in Physiology or Medicine for their work on RNAi in the nematode worm '' Caenorhabditis elegans'', which they published in 1998. Since the discovery of RNAi and its regulatory potentials, it has become evident that RNAi has immense potential in suppression of desired genes. RNAi is now known as precise, efficient, stable and better than antisense therapy for gene suppression. Antisense RNA produced intracellularly by an expression vector m ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Gut Flora
Gut microbiota, gut microbiome, or gut flora, are the microorganisms, including bacteria, archaea, fungi, and viruses that live in the digestive tracts of animals. The gastrointestinal metagenome is the aggregate of all the genomes of the gut microbiota. The gut is the main location of the human microbiome. The gut microbiota has broad impacts, including effects on colonization, resistance to pathogens, maintaining the intestinal epithelium, metabolizing dietary and pharmaceutical compounds, controlling immune function, and even behavior through the gut–brain axis. The microbial composition of the gut microbiota varies across regions of the digestive tract. The colon contains the highest microbial density recorded in any habitat on Earth, representing between 300 and 1000 different species. Bacteria are the largest and to date, best studied component and 99% of gut bacteria come from about 30 or 40 species. Up to 60% of the dry mass of feces is bacteria. Over 99% of the bac ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |