|
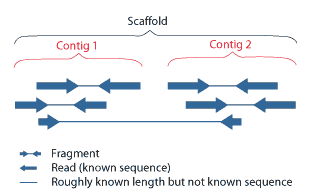
Contig Map
A contig (from ''contiguous'') is a set of overlapping DNA segments that together represent a consensus region of DNA.Gregory, S. ''Contig Assembly''. Encyclopedia of Life Sciences, 2005. In bottom-up sequencing projects, a contig refers to overlapping sequence data ( reads); in top-down sequencing projects, contig refers to the overlapping clones that form a physical map of the genome that is used to guide sequencing and assembly.Dear, P. H. ''Genome Mapping''. Encyclopedia of Life Sciences, 2005. . Contigs can thus refer both to overlapping DNA sequences and to overlapping physical segments (fragments) contained in clones depending on the context. Original definition of contig In 1980, Staden wrote: ''In order to make it easier to talk about our data gained by the shotgun method of sequencing we have invented the word "contig". A contig is a set of gel readings that are related to one another by overlap of their sequences. All gel readings belong to one and only one con ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Consensus Sequence
In molecular biology and bioinformatics, the consensus sequence (or canonical sequence) is the calculated order of most frequent residues, either nucleotide or amino acid, found at each position in a sequence alignment. It serves as a simplified representation of the population. It represents the results of multiple sequence alignments in which related sequences are compared to each other and similar sequence motifs are calculated. Such information is important when considering sequence-dependent enzymes such as RNA polymerase.Pierce, Benjamin A. 2002. Genetics : A Conceptual Approach. 1st ed. New York: W.H. Freeman and Co. Biological significance A protein binding site, represented by a consensus sequence, may be a short sequence of nucleotides which is found several times in the genome and is thought to play the same role in its different locations. For example, many transcription factors recognize particular patterns in the promoters of the genes they regulate. In the same way, ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Bacterial Artificial Chromosome
A bacterial artificial chromosome (BAC) is a DNA construct, based on a functional fertility plasmid (or F-plasmid), used for transforming and cloning in bacteria, usually '' E. coli''. F-plasmids play a crucial role because they contain partition genes that promote the even distribution of plasmids after bacterial cell division. The bacterial artificial chromosome's usual insert size is 150–350 kbp. A similar cloning vector called a PAC has also been produced from the DNA of P1 bacteriophage. BACs are often used to sequence the genome of organisms in genome projects, for example the Human Genome Project. A short piece of the organism's DNA is amplified as an insert in BACs, and then sequenced. Finally, the sequenced parts are rearranged ''in silico'', resulting in the genomic sequence of the organism. BACs were replaced with faster and less laborious sequencing methods like whole genome shotgun sequencing and now more recently next-gen sequencing. Common gene components ;''re ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Staden Package
The Staden Package is computer software, a set of tools for DNA sequence assembly, editing, and sequence analysis. It is open-source software, released under a BSD 3-clause license. Package components The Staden package consists of several different programs. The main components are: * pregap4 – base calling with Phred, end clipping, and vector trimming * trev – trace viewing and editing * gap4 – sequence assembly, contig editing, and finishing * gap5 – assembly visualising, editing, and finishing of NGS data * Spin – DNA and protein sequence analysis History The Staden Package was developed by Rodger Staden's group at the Medical Research Council (MRC) Laboratory of Molecular Biology, Cambridge, England, since 1977. The package was available free to academic users, with 2,500 licenses issued in 2003 and an estimated 10,000 users, when funding for further development ended. The package was converted to open-source in 2004, and several new versions have been rele ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Primer Walking
Primer walking is a technique used to clone a gene (e.g., disease gene) from its known closest markers (e.g., known gene). As a result, it is employed in cloning and sequencing efforts in plants, fungi, and mammals with minor alterations. This technique, also known as "directed sequencing," employs a series of Sanger sequencing reactions to either confirm the reference sequence of a known plasmid or PCR product based on the reference sequence (sequence confirmation service) or to discover the unknown sequence of a full plasmid or PCR product by designing primers to sequence overlapping sections (sequence discovery service). Primer walking: a DNA sequencing method Primer walking is a method to determine the sequence of DNA up to the 1.3–7.0 kb range whereas chromosome walking is used to produce the clones of already known sequences of the gene. Too long fragments cannot be sequenced in a single sequence read using the chain termination method. This method works by dividing the ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Gel Electrophoresis
Gel electrophoresis is a method for separation and analysis of biomacromolecules ( DNA, RNA, proteins, etc.) and their fragments, based on their size and charge. It is used in clinical chemistry to separate proteins by charge or size (IEF agarose, essentially size independent) and in biochemistry and molecular biology to separate a mixed population of DNA and RNA fragments by length, to estimate the size of DNA and RNA fragments or to separate proteins by charge. Nucleic acid molecules are separated by applying an electric field to move the negatively charged molecules through a matrix of agarose or other substances. Shorter molecules move faster and migrate farther than longer ones because shorter molecules migrate more easily through the pores of the gel. This phenomenon is called sieving. Proteins are separated by the charge in agarose because the pores of the gel are too small to sieve proteins. Gel electrophoresis can also be used for the separation of nanoparticles. ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Restriction Enzyme
A restriction enzyme, restriction endonuclease, REase, ENase or'' restrictase '' is an enzyme that cleaves DNA into fragments at or near specific recognition sites within molecules known as restriction sites. Restriction enzymes are one class of the broader endonuclease group of enzymes. Restriction enzymes are commonly classified into five types, which differ in their structure and whether they cut their DNA substrate at their recognition site, or if the recognition and cleavage sites are separate from one another. To cut DNA, all restriction enzymes make two incisions, once through each sugar-phosphate backbone (i.e. each strand) of the DNA double helix. These enzymes are found in bacteria and archaea and provide a defense mechanism against invading viruses. Inside a prokaryote, the restriction enzymes selectively cut up ''foreign'' DNA in a process called ''restriction digestion''; meanwhile, host DNA is protected by a modification enzyme (a methyltransferase) that modifi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Restriction Digest
A restriction digest is a procedure used in molecular biology to prepare DNA for analysis or other processing. It is sometimes termed ''DNA fragmentation'' (this term is used for other procedures as well). Hartl and Jones describe it this way: This enzymatic technique can be used for cleaving DNA molecules at specific sites, ensuring that all DNA fragments that contain a particular sequence at a particular location have the same size; furthermore, each fragment that contains the desired sequence has the sequence located at exactly the same position within the fragment. The cleavage method makes use of an important class of DNA-cleaving enzymes isolated primarily from bacteria. These enzymes are called restriction endonucleases or restriction enzymes, and they are able to cleave DNA molecules at the positions at which particular short sequences of bases are present.Hartl, Daniel L., Jones, Elizabeth W. (2001), ''Genetics: Analysis of Genes and Genomes'', Fifth Edition. The res ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Sequence-tagged Site
A sequence-tagged site (or STS) is a short (200 to 500 base pair) DNA sequence that has a single occurrence in the genome and whose location and base sequence are known. Usage STSs can be easily detected by the polymerase chain reaction (PCR) using specific primers. For this reason they are useful for constructing genetic and physical maps from sequence data reported from many different laboratories. They serve as landmarks on the developing physical map of a genome. When STS loci contain genetic polymorphisms (e.g. simple sequence length polymorphisms, SSLPs, single nucleotide polymorphisms), they become valuable genetic markers, i.e. loci which can be used to distinguish individuals. They are used in shotgun sequencing, specifically to aid sequence assembly. STSs are very helpful for detecting microdeletions in some genes. For example, some STSs can be used in screening by PCR to detect microdeletions in Azoospermia (AZF) genes in infertile men. References External links ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Library (biology)
In molecular biology, a library is a collection of DNA fragments that is stored and propagated in a population of micro-organisms through the process of molecular cloning. There are different types of DNA libraries, including cDNA libraries (formed from reverse-transcribed RNA), genomic libraries (formed from genomic DNA) and randomized mutant libraries (formed by de novo gene synthesis where alternative nucleotides or codons are incorporated). DNA library technology is a mainstay of current molecular biology, genetic engineering, and protein engineering, and the applications of these libraries depend on the source of the original DNA fragments. There are differences in the cloning vectors and techniques used in library preparation, but in general each DNA fragment is uniquely inserted into a cloning vector and the pool of recombinant DNA molecules is then transferred into a population of bacteria (a Bacterial Artificial Chromosome or BAC library) or yeast such that each or ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
P1-derived Artificial Chromosome
A P1-derived artificial chromosome, or PAC, is a DNA construct derived from the DNA of P1 bacteriophages and Bacterial artificial chromosome. It can carry large amounts (about 100–300 kilobases) of other sequences for a variety of bioengineering purposes in bacteria. It is one type of the efficient cloning vector used to clone DNA fragments (100- to 300-kb insert size; average,150 kb) in ''Escherichia coli'' cells. History of PAC The bacteriophage P1 was first isolated by Dr. Giuseppe Bertani. In his study, he noticed that the lysogen produced abnormal non-continuous phages, and later found phage P1 was produced from the Lisbonne lysogen strain, in addition to bacteriophages P2 and P3. P1 has the ability to copy a bacteria's host genome and integrate that DNA information into other bacteria hosts, also known as generalized transduction. Later on, P1 was developed as a cloning vector by Nat Sternberg and colleagues in the 1990s. It is capable of Cre-Lox recombination. The P1 ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Bacterial Artificial Chromosome
A bacterial artificial chromosome (BAC) is a DNA construct, based on a functional fertility plasmid (or F-plasmid), used for transforming and cloning in bacteria, usually '' E. coli''. F-plasmids play a crucial role because they contain partition genes that promote the even distribution of plasmids after bacterial cell division. The bacterial artificial chromosome's usual insert size is 150–350 kbp. A similar cloning vector called a PAC has also been produced from the DNA of P1 bacteriophage. BACs are often used to sequence the genome of organisms in genome projects, for example the Human Genome Project. A short piece of the organism's DNA is amplified as an insert in BACs, and then sequenced. Finally, the sequenced parts are rearranged ''in silico'', resulting in the genomic sequence of the organism. BACs were replaced with faster and less laborious sequencing methods like whole genome shotgun sequencing and now more recently next-gen sequencing. Common gene components ;''re ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Mate Pair
Paired-end tags (PET) (sometimes "Paired-End diTags", or simply "ditags") are the short sequences at the 5’ and 3' ends of a DNA fragment which are unique enough that they (theoretically) exist together only once in a genome, therefore making the sequence of the DNA in between them available upon search (if full-genome sequence data is available) or upon further sequencing (since tag sites are unique enough to serve as primer annealing sites). Paired-end tags (PET) exist in PET libraries with the intervening DNA absent, that is, a PET "represents" a larger fragment of genomic or cDNA by consisting of a short 5' linker sequence, a short 5' sequence tag, a short 3' sequence tag, and a short 3' linker sequence. It was shown conceptually that 13 base pairs are sufficient to map tags uniquely.Fullwood MJ, Wei CL, Liu ET, Ruan Y. 2009. Next-Generation DNA sequencing of paired-end tags (PET) for transcriptome and genome analyses. Genome Research. 19:521–532. {{PMID, 19339662 However ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |