|
Library (biology)
In molecular biology, a library is a collection of DNA fragments that is stored and propagated in a population of micro-organisms through the process of molecular cloning. There are different types of DNA libraries, including cDNA libraries (formed from reverse-transcribed RNA), genomic libraries (formed from genomic DNA) and randomized mutant libraries (formed by de novo gene synthesis where alternative nucleotides or codons are incorporated). DNA library technology is a mainstay of current molecular biology, genetic engineering, and protein engineering, and the applications of these libraries depend on the source of the original DNA fragments. There are differences in the cloning vectors and techniques used in library preparation, but in general each DNA fragment is uniquely inserted into a cloning vector and the pool of recombinant DNA molecules is then transferred into a population of bacteria (a Bacterial Artificial Chromosome or BAC library) or yeast such that each o ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
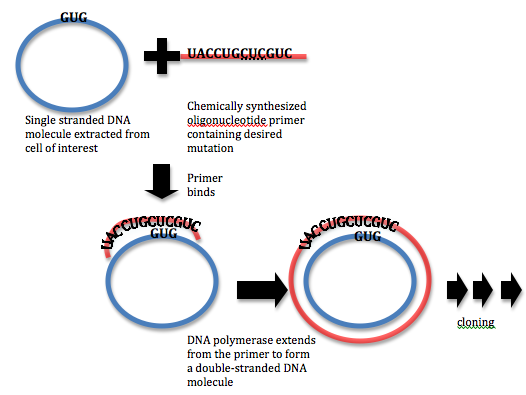
Site Saturation Mutagenesis
Site saturation mutagenesis (SSM), or simply site saturation, is a Mutagenesis (molecular biology technique), random mutagenesis technique used in protein engineering, in which a single codon or set of codons is substituted with all possible amino acids at the position. There are many variants of the site saturation technique, from paired site saturation (saturating two positions in every mutant in the library) to scanning site saturation (performing a site saturation at every site in the protein, resulting in a library of size [20 x (number of residues in the protein)] that contains every possible point mutation, point mutant of the protein). Method Saturation mutagenesis is commonly achieved by site-directed mutagenesis PCR with a randomised codon in the primers (e.g. SeSaM) or by Artificial gene synthesis#Applications, artificial gene synthesis, with a mixture of synthesis nucleotides used at the codons to be randomised. Different degenerate codons can be used to encode set ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Genomic Library
A genomic library is a collection of the total genomic DNA from a single organism. The DNA is stored in a population of identical vectors, each containing a different insert of DNA. In order to construct a genomic library, the organism's DNA is extracted from cells and then digested with a restriction enzyme to cut the DNA into fragments of a specific size. The fragments are then inserted into the vector using DNA ligase. Next, the vector DNA can be taken up by a host organism - commonly a population of Escherichia coli or yeast - with each cell containing only one vector molecule. Using a host cell to carry the vector allows for easy amplification and retrieval of specific clones from the library for analysis. There are several kinds of vectors available with various insert capacities. Generally, libraries made from organisms with larger genomes require vectors featuring larger inserts, thereby fewer vector molecules are needed to make the library. Researchers can choose a vec ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
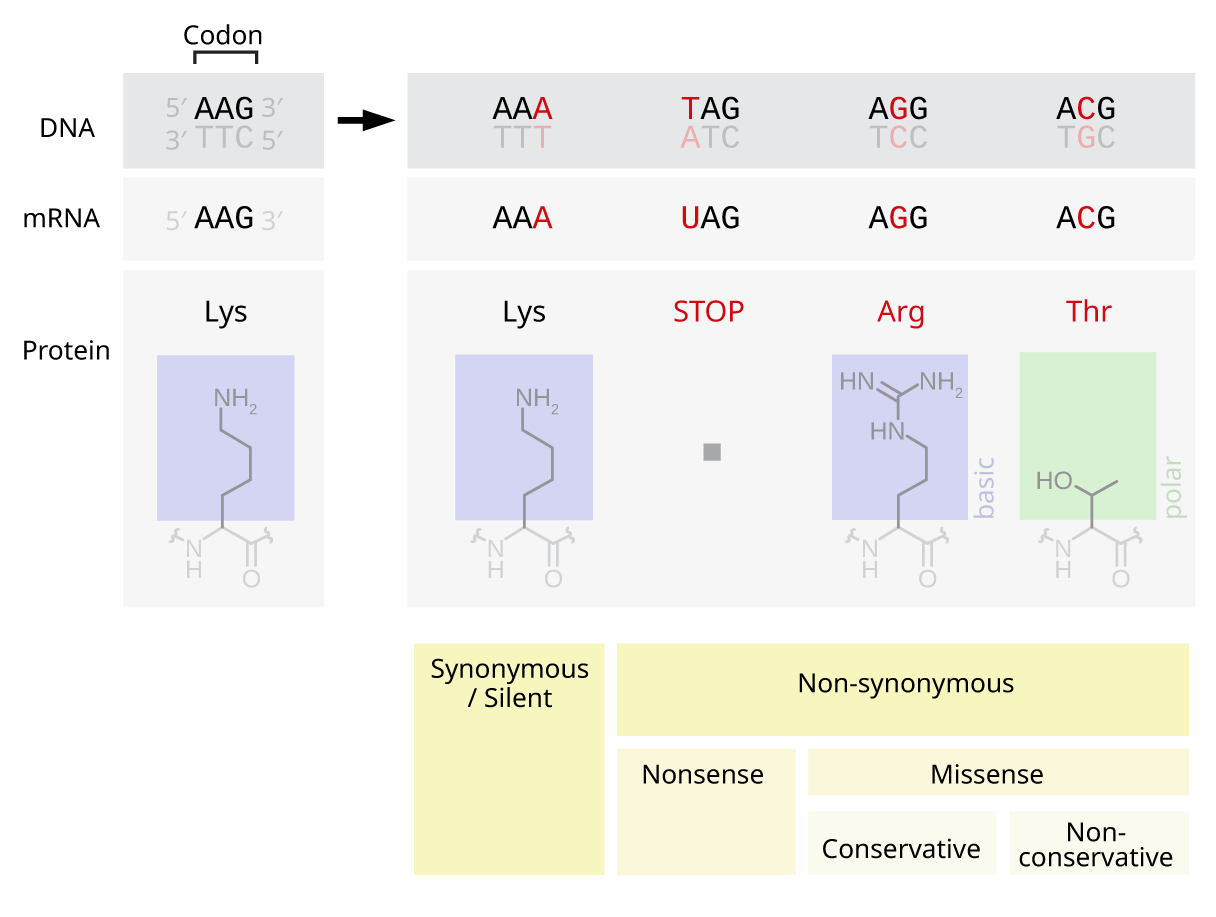
Point Mutation
A point mutation is a genetic mutation where a single nucleotide base is changed, inserted or deleted from a DNA or RNA sequence of an organism's genome. Point mutations have a variety of effects on the downstream protein product—consequences that are moderately predictable based upon the specifics of the mutation. These consequences can range from no effect (e.g. synonymous mutations) to deleterious effects (e.g. frameshift mutations), with regard to protein production, composition, and function. Causes Point mutations usually take place during DNA replication. DNA replication occurs when one double-stranded DNA molecule creates two single strands of DNA, each of which is a template for the creation of the complementary strand. A single point mutation can change the whole DNA sequence. Changing one purine or pyrimidine may change the amino acid that the nucleotides code for. Point mutations may arise from spontaneous mutations that occur during DNA replication. The rate of ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Saturation Mutagenesis
Site saturation mutagenesis (SSM), or simply site saturation, is a random mutagenesis technique used in protein engineering, in which a single codon or set of codons is substituted with all possible amino acids at the position. There are many variants of the site saturation technique, from paired site saturation (saturating two positions in every mutant in the library) to scanning site saturation (performing a site saturation at every site in the protein, resulting in a library of size 0 x (number of residues in the protein)that contains every possible point mutant of the protein). Method Saturation mutagenesis is commonly achieved by site-directed mutagenesis PCR with a randomised codon in the primers (e.g. SeSaM) or by artificial gene synthesis, with a mixture of synthesis nucleotide Nucleotides are organic molecules consisting of a nucleoside and a phosphate. They serve as monomeric units of the nucleic acid polymers – deoxyribonucleic acid (DNA) and ribon ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Artificial Gene Synthesis
Artificial gene synthesis, or simply gene synthesis, refers to a group of methods that are used in synthetic biology to construct and assemble genes from nucleotides '' de novo''. Unlike DNA synthesis in living cells, artificial gene synthesis does not require template DNA, allowing virtually any DNA sequence to be synthesized in the laboratory. It comprises two main steps, the first of which is solid-phase DNA synthesis, sometimes known as DNA printing. This produces oligonucleotide fragments that are generally under 200 base pairs. The second step then involves connecting these oligonucleotide fragments using various DNA assembly methods. Because artificial gene synthesis does not require template DNA, it is theoretically possible to make a completely synthetic DNA molecule with no limits on the nucleotide sequence or size. Synthesis of the first complete gene, a yeast tRNA, was demonstrated by Har Gobind Khorana and coworkers in 1972. Synthesis of the first peptide- and ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Indel
Indel is a molecular biology term for an insertion or deletion of bases in the genome of an organism. It is classified among small genetic variations, measuring from 1 to 10 000 base pairs in length, including insertion and deletion events that may be separated by many years, and may not be related to each other in any way. A microindel is defined as an indel that results in a net change of 1 to 50 nucleotides. In coding regions of the genome, unless the length of an indel is a multiple of 3, it will produce a frameshift mutation. For example, a common microindel which results in a frameshift causes Bloom syndrome in the Jewish or Japanese population. Indels can be contrasted with a point mutation. An indel inserts or deletes nucleotides from a sequence, while a point mutation is a form of substitution that ''replaces'' one of the nucleotides without changing the overall number in the DNA. Indels can also be contrasted with Tandem Base Mutations (TBM), which may result from fu ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
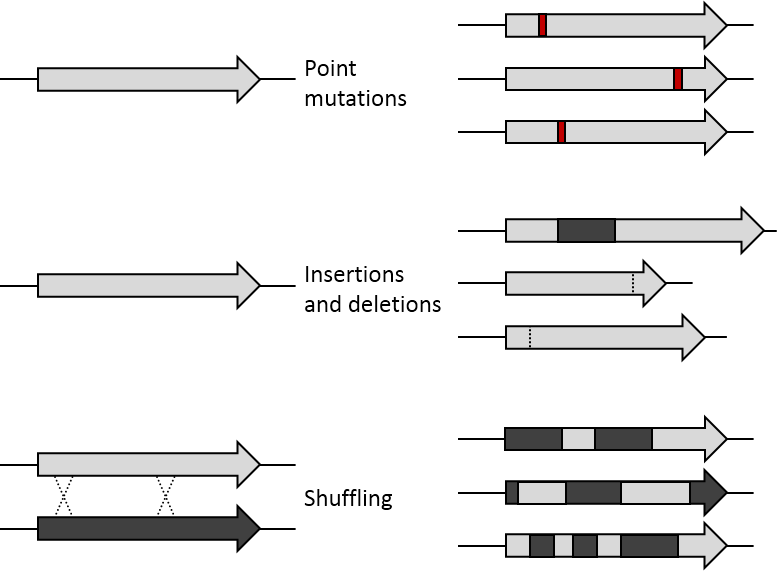
DNA Shuffling
DNA shuffling, also known as molecular breeding, is an in vitro random recombination method to generate mutant genes for directed evolution and to enable a rapid increase in DNA library size. Three procedures for accomplishing DNA shuffling are molecular breeding which relies on homologous recombination or the similarity of the DNA sequences, restriction enzymes which rely on common restriction sites, and nonhomologous random recombination which requires the use of hairpins. In all of these techniques, the parent genes are fragmented and then recombined. DNA shuffling utilizes random recombination as opposed to site-directed mutagenesis in order to generate proteins with unique attributes or combinations of desirable characteristics encoded in the parent genes such as thermostability and high activity. The potential for DNA shuffling to produce novel proteins is exemplified by the figure shown on the right which demonstrates the difference between point mutations, insertions ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Error-prone PCR
In molecular biology, mutagenesis is an important laboratory technique whereby DNA mutations are deliberately engineered to produce libraries of mutant genes, proteins, strains of bacteria, or other genetically modified organisms. The various constituents of a gene, as well as its regulatory elements and its gene products, may be mutated so that the functioning of a genetic locus, process, or product can be examined in detail. The mutation may produce mutant proteins with interesting properties or enhanced or novel functions that may be of commercial use. Mutant strains may also be produced that have practical application or allow the molecular basis of a particular cell function to be investigated. Many methods of mutagenesis exist today. Initially, the kind of mutations artificially induced in the laboratory were entirely random using mechanisms such as UV irradiation. Random mutagenesis cannot target specific regions or sequences of the genome; however, with the development ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cancer
Cancer is a group of diseases involving abnormal cell growth with the potential to invade or spread to other parts of the body. These contrast with benign tumors, which do not spread. Possible signs and symptoms include a lump, abnormal bleeding, prolonged cough, unexplained weight loss, and a change in bowel movements. While these symptoms may indicate cancer, they can also have other causes. Over 100 types of cancers affect humans. Tobacco use is the cause of about 22% of cancer deaths. Another 10% are due to obesity, poor diet, lack of physical activity or excessive drinking of alcohol. Other factors include certain infections, exposure to ionizing radiation, and environmental pollutants. In the developing world, 15% of cancers are due to infections such as '' Helicobacter pylori'', hepatitis B, hepatitis C, human papillomavirus infection, Epstein–Barr virus and human immunodeficiency virus (HIV). These factors act, at least partly, by changing the genes o ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Genetic Mutation
In biology, a mutation is an alteration in the nucleic acid sequence of the genome of an organism, virus, or extrachromosomal DNA. Viral genomes contain either DNA or RNA. Mutations result from errors during DNA or viral replication, mitosis, or meiosis or other types of damage to DNA (such as pyrimidine dimers caused by exposure to ultraviolet radiation), which then may undergo error-prone repair (especially microhomology-mediated end joining), cause an error during other forms of repair, or cause an error during replication ( translesion synthesis). Mutations may also result from insertion or deletion of segments of DNA due to mobile genetic elements. Mutations may or may not produce detectable changes in the observable characteristics (phenotype) of an organism. Mutations play a part in both normal and abnormal biological processes including: evolution, cancer, and the development of the immune system, including junctional diversity. Mutation is the ultimate source of ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Regulatory Sequences
A regulatory sequence is a segment of a nucleic acid molecule which is capable of increasing or decreasing the expression of specific genes within an organism. Regulation of gene expression is an essential feature of all living organisms and viruses. Description In DNA, regulation of gene expression normally happens at the level of RNA biosynthesis ( transcription). It is accomplished through the sequence-specific binding of proteins (transcription factors) that activate or inhibit transcription. Transcription factors may act as activators, repressors, or both. Repressors often act by preventing RNA polymerase from forming a productive complex with the transcriptional initiation region ( promoter), while activators facilitate formation of a productive complex. Furthermore, DNA motifs have been shown to be predictive of epigenomic modifications, suggesting that transcription factors play a role in regulating the epigenome. In RNA, regulation may occur at the level of protei ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |