|
Candidate Phyla
A candidate division, candidate phylum or candidate division-level is a lineage of prokaryotic organisms for which until recently no cultured representatives have been found, but evidence of the existence of the clade has been obtained by 16S rRNA and metagenomic analysis of environmental samples. The term candidatus in fact means a species for which there is insufficient information to call it a new species according to the International Code of Nomenclature of Bacteria Examples of Candidate division bacteria * Candidate division TM7 (Saccharibacteria) * Poribacteria * TG3 (candidate phylum) (Chitinivibrionia) See also * Metagenomics * Cloning * DNA sequencing DNA sequencing is the process of determining the nucleic acid sequence – the order of nucleotides in DNA. It includes any method or technology that is used to determine the order of the four bases: adenine, guanine, cytosine, and thymine. Th ... References {{reflist Candidatus taxa Bacteria phyla Archae ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Prokaryote
A prokaryote () is a single-celled organism that lacks a nucleus and other membrane-bound organelles. The word ''prokaryote'' comes from the Greek πρό (, 'before') and κάρυον (, 'nut' or 'kernel').Campbell, N. "Biology:Concepts & Connections". Pearson Education. San Francisco: 2003. In the two-empire system arising from the work of Édouard Chatton, prokaryotes were classified within the empire Prokaryota. But in the three-domain system, based upon molecular analysis, prokaryotes are divided into two domains: ''Bacteria'' (formerly Eubacteria) and ''Archaea'' (formerly Archaebacteria). Organisms with nuclei are placed in a third domain, Eukaryota. In the study of the origins of life, prokaryotes are thought to have arisen before eukaryotes. Besides the absence of a nucleus, prokaryotes also lack mitochondria, or most of the other membrane-bound organelles that characterize the eukaryotic cell. It was once thought that prokaryotic cellular components within the cytop ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Microbiology
Microbiology () is the scientific study of microorganisms, those being unicellular (single cell), multicellular (cell colony), or acellular (lacking cells). Microbiology encompasses numerous sub-disciplines including virology, bacteriology, protistology, mycology, immunology, and parasitology. Eukaryotic microorganisms possess membrane-bound organelles and include fungi and protists, whereas prokaryotic organisms—all of which are microorganisms—are conventionally classified as lacking membrane-bound organelles and include Bacteria and Archaea. Microbiologists traditionally relied on culture, staining, and microscopy. However, less than 1% of the microorganisms present in common environments can be cultured in isolation using current means. Microbiologists often rely on molecular biology tools such as DNA sequence based identification, for example the 16S rRNA gene sequence used for bacteria identification. Viruses have been variably classified as organisms, as they have ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
16S Ribosomal RNA
16 S ribosomal RNA (or 16 S rRNA) is the RNA component of the 30S subunit of a prokaryotic ribosome (SSU rRNA). It binds to the Shine-Dalgarno sequence and provides most of the SSU structure. The genes coding for it are referred to as 16S rRNA gene and are used in reconstructing phylogenies, due to the slow rates of evolution of this region of the gene. Carl Woese and George E. Fox were two of the people who pioneered the use of 16S rRNA in phylogenetics in 1977. Multiple sequences of the 16S rRNA gene can exist within a single bacterium. Functions * Like the large (23S) ribosomal RNA, it has a structural role, acting as a scaffold defining the positions of the ribosomal proteins. * The 3-end contains the anti- Shine-Dalgarno sequence, which binds upstream to the AUG start codon on the mRNA. The 3-end of 16S RNA binds to the proteins S1 and S21 which are known to be involved in initiation of protein synthesis * Interacts with 23S, aiding in the binding of the two ribosomal s ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
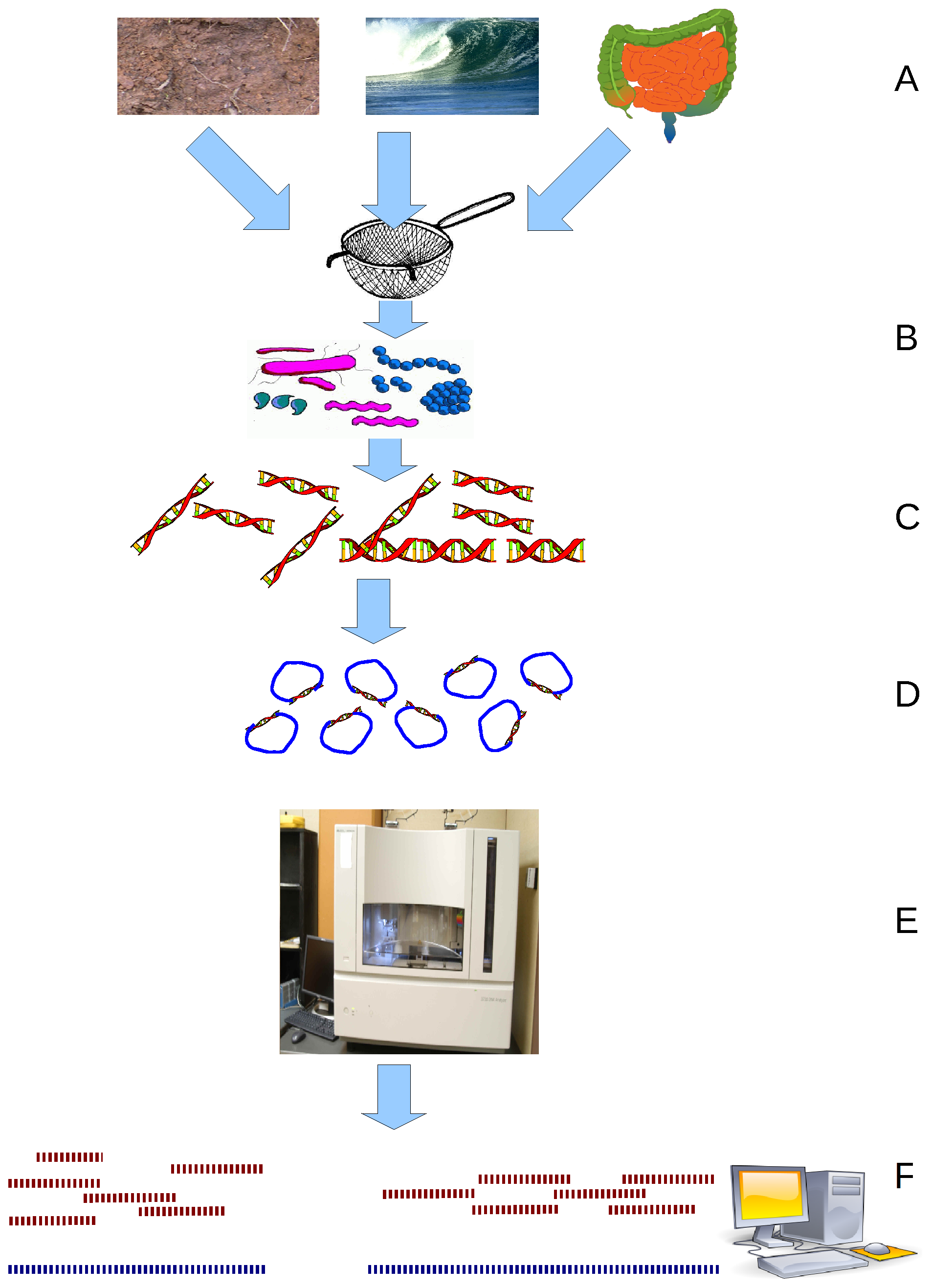
Metagenomics
Metagenomics is the study of genetic material recovered directly from environmental or clinical samples by a method called sequencing. The broad field may also be referred to as environmental genomics, ecogenomics, community genomics or microbiomics. While traditional microbiology and microbial genome sequencing and genomics rely upon cultivated clonal cultures, early environmental gene sequencing cloned specific genes (often the 16S rRNA gene) to produce a profile of diversity in a natural sample. Such work revealed that the vast majority of microbial biodiversity had been missed by cultivation-based methods. Because of its ability to reveal the previously hidden diversity of microscopic life, metagenomics offers a powerful lens for viewing the microbial world that has the potential to revolutionize understanding of the entire living world. As the price of DNA sequencing continues to fall, metagenomics now allows microbial ecology to be investigated at a much greater scale ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Candidatus
In prokaryote nomenclature, ''Candidatus'' (Latin for candidate of Roman office) is used to name prokaryotic phyla that are well characterized but yet-uncultured. Contemporary sequencing approaches, such as 16S sequencing or metagenomics, provide much information about the analyzed organisms and thus allow to identify and characterize individual species. However, the majority of prokaryotic species remain uncultivable and hence inaccessible for further characterization in ''in vitro'' study. The recent discoveries of a multitude of candidate taxa has led to candidate phyla radiation expanding the tree of life through the new insights in bacterial diversity. Nomenclature History The initial International Code of Nomenclature of Prokaryotes as well as early revisions did not account for the possibility of identifying prokaryotes which were not yet cultivable. Therefore, the term ''Candidatus'' was proposed in the context of a conference of the International Committee on Systemati ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
International Code Of Nomenclature Of Bacteria
The International Code of Nomenclature of Prokaryotes (ICNP) formerly the International Code of Nomenclature of Bacteria (ICNB) or Bacteriological Code (BC) governs the scientific names for Bacteria and Archaea.P. H. A. Sneath, 2003. A short history of the Bacteriological CodURL It denotes the rules for naming taxa of bacteria, according to their relative rank. As such it is one of the nomenclature codes of biology. Originally the ''International Code of Botanical Nomenclature'' dealt with bacteria, and this kept references to bacteria until these were eliminated at the 1975 International Botanical Congress. An early Code for the nomenclature of bacteria was approved at the 4th International Congress for Microbiology in 1947, but was later discarded. The latest version to be printed in book form is the 1990 Revision, but the book does not represent the current rules. The 2008 Revision has been published in the ''International Journal of Systematic and Evolutionary Microbiology'' (I ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Saccharibacteria
Saccharibacteria, formerly known as ''TM7'', is a major bacterial lineage. It was discovered through 16S rRNA sequencing . TM7x from the human oral cavity was cultivated and revealed that TM7x is an extremely small coccus (200-300 nm) and has a distinctive lifestyle not previously observed in human-associated microbes. It is an obligate epibiont of various hosts, including ''Actinomyces odontolyticus'' strain (XH001) yet also has a parasitic phase thereby killing its host. The full genome sequence revealed a highly reduced genome (705kB) and a complete lack of amino acid biosynthetic capacity. An axenic culture of TM7 from the oral cavity was reported in 2014 but no sequence or culture was made available. Along with Candidate Phylum TM6, it was named after sequences obtained in 1994 in an environmental study of a soil sample of peat bog in Germany where 262 PCR amplified 16S rDNA fragments were cloned into a plasmid vector, named TM clones for ' ( lit. peat, middle lay ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Poribacteria
Poribacteria are a candidate phylum of bacteria originally discovered in the microbiome of marine sponges (''Porifera''). Poribacteria are Gram-negative primarily aerobic mixotrophs with the ability for oxidative phosphorylation, glycolysis, and autotrophic carbon fixation via the Wood – Ljungdahl pathway. Poribacterial heterotrophy is characterised by an enriched set of glycoside hydrolases, uronic acid degradation, as well as several specific sulfatases. This heterotrophic repertoire of poribacteria was suggested to be involved in the degradation of the extracellular sponge host matrix. Genome Single-cell genomics and metagenomic shotgun sequencing approaches reveal a poribacterial genome size range between about 4.2 and 6.5 megabases encoding 4,254 protein-coding genes, of which an unusually high 24% have no homology to known genes. Among the genes of identifiable homology, reconstructed pathways suggest that the poribacterial central metabolism is capable of glycolysis, tr ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
TG3 (candidate Phylum)
Candidate phylum TG3 (aka candidate division Termite Group 3 or Chitinivibrionia Sorokin et al. 2014)NCBIChitinivibrionia Sorokin et al. 2014(class, syn. candidate division TG3) is a candidate phylum that is closely related to the phylum ''Fibrobacterota'' based on 16S rRNA gene phylogeny. Originally thought to be composed solely of sequences from termite Termites are small insects that live in colonies and have distinct castes (eusocial) and feed on wood or other dead plant matter. Termites comprise the infraorder Isoptera, or alternatively the epifamily Termitoidae, within the order Blattode ... guts, it was later found to be more widespread in nature. References {{Taxonbar, from=Q7670148 Bacteria phyla Termites Candidatus taxa ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Molecular Cloning
Molecular cloning is a set of experimental methods in molecular biology that are used to assemble recombinant DNA molecules and to direct their replication within host organisms. The use of the word ''cloning'' refers to the fact that the method involves the replication of one molecule to produce a population of cells with identical DNA molecules. Molecular cloning generally uses DNA sequences from two different organisms: the species that is the source of the DNA to be cloned, and the species that will serve as the living host for replication of the recombinant DNA. Molecular cloning methods are central to many contemporary areas of modern biology and medicine. In a conventional molecular cloning experiment, the DNA to be cloned is obtained from an organism of interest, then treated with enzymes in the test tube to generate smaller DNA fragments. Subsequently, these fragments are then combined with vector DNA to generate recombinant DNA molecules. The recombinant DNA is then in ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DNA Sequencing
DNA sequencing is the process of determining the nucleic acid sequence – the order of nucleotides in DNA. It includes any method or technology that is used to determine the order of the four bases: adenine, guanine, cytosine, and thymine. The advent of rapid DNA sequencing methods has greatly accelerated biological and medical research and discovery. Knowledge of DNA sequences has become indispensable for basic biological research, DNA Genographic Projects and in numerous applied fields such as medical diagnosis, biotechnology, forensic biology, virology and biological systematics. Comparing healthy and mutated DNA sequences can diagnose different diseases including various cancers, characterize antibody repertoire, and can be used to guide patient treatment. Having a quick way to sequence DNA allows for faster and more individualized medical care to be administered, and for more organisms to be identified and cataloged. The rapid speed of sequencing attained with modern D ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Candidatus Taxa
In prokaryote nomenclature, ''Candidatus'' (Latin for candidate of Roman office) is used to name prokaryotic phyla that are well characterized but yet-uncultured. Contemporary sequencing approaches, such as 16S sequencing or metagenomics, provide much information about the analyzed organisms and thus allow to identify and characterize individual species. However, the majority of prokaryotic species remain uncultivable and hence inaccessible for further characterization in ''in vitro'' study. The recent discoveries of a multitude of candidate taxa has led to candidate phyla radiation expanding the tree of life through the new insights in bacterial diversity. Nomenclature History The initial International Code of Nomenclature of Prokaryotes as well as early revisions did not account for the possibility of identifying prokaryotes which were not yet cultivable. Therefore, the term ''Candidatus'' was proposed in the context of a conference of the International Committee on Systemati ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |