|
Cancer Genome Sequencing
Cancer genome sequencing is the whole genome sequencing of a single, homogeneous or heterogeneous group of cancer cells. It is a biochemical laboratory method for the characterization and identification of the DNA or RNA sequences of cancer cell(s). Unlike whole genome (WG) sequencing which is typically from blood cells, such as J. Craig Venter's and James D. Watson’s WG sequencing projects, saliva, epithelial cells or bone - cancer genome sequencing involves direct sequencing of primary tumor tissue, adjacent or distal normal tissue, the tumor micro environment such as fibroblast/stromal cells, or metastatic tumor sites. Similar to whole genome sequencing, the information generated from this technique include: identification of nucleotide bases (DNA or RNA), copy number and sequence variants, mutation status, and structural changes such as chromosomal translocations and fusion genes. Cancer genome sequencing is not limited to WG sequencing and can also include exome, transcript ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Whole Genome Sequencing
Whole genome sequencing (WGS), also known as full genome sequencing, complete genome sequencing, or entire genome sequencing, is the process of determining the entirety, or nearly the entirety, of the DNA sequence of an organism's genome at a single time. This entails sequencing all of an organism's chromosomal DNA as well as DNA contained in the mitochondrial DNA, mitochondria and, for plants, in the chloroplast. Whole genome sequencing has largely been used as a research tool, but was being introduced to clinics in 2014. In the future of personalized medicine, whole genome sequence data may be an important tool to guide therapeutic intervention. The tool of DNA sequencing, gene sequencing at Single-nucleotide polymorphism, SNP level is also used to pinpoint functional variants from association studies and improve the knowledge available to researchers interested in evolutionary biology, and hence may lay the foundation for predicting disease susceptibility and drug response. ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Epigenetic
In biology, epigenetics is the study of stable phenotypic changes (known as ''marks'') that do not involve alterations in the DNA sequence. The Greek prefix '' epi-'' ( "over, outside of, around") in ''epigenetics'' implies features that are "on top of" or "in addition to" the traditional genetic basis for inheritance. Epigenetics most often involves changes that affect the regulation of gene expression, but the term can also be used to describe any heritable phenotypic change. Such effects on cellular and physiological phenotypic traits may result from external or environmental factors, or be part of normal development. The term also refers to the mechanism of changes: functionally relevant alterations to the genome that do not involve mutation of the nucleotide sequence. Examples of mechanisms that produce such changes are DNA methylation and histone modification, each of which alters how genes are expressed without altering the underlying DNA sequence. Gene expression can ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Genome Assembly
In bioinformatics, sequence assembly refers to aligning and merging fragments from a longer DNA sequence in order to reconstruct the original sequence. This is needed as DNA sequencing technology might not be able to 'read' whole genomes in one go, but rather reads small pieces of between 20 and 30,000 bases, depending on the technology used. Typically, the short fragments (reads) result from shotgun sequencing genomic DNA, or gene transcript ( ESTs). The problem of sequence assembly can be compared to taking many copies of a book, passing each of them through a shredder with a different cutter, and piecing the text of the book back together just by looking at the shredded pieces. Besides the obvious difficulty of this task, there are some extra practical issues: the original may have many repeated paragraphs, and some shreds may be modified during shredding to have typos. Excerpts from another book may also be added in, and some shreds may be completely unrecognizable. Gen ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
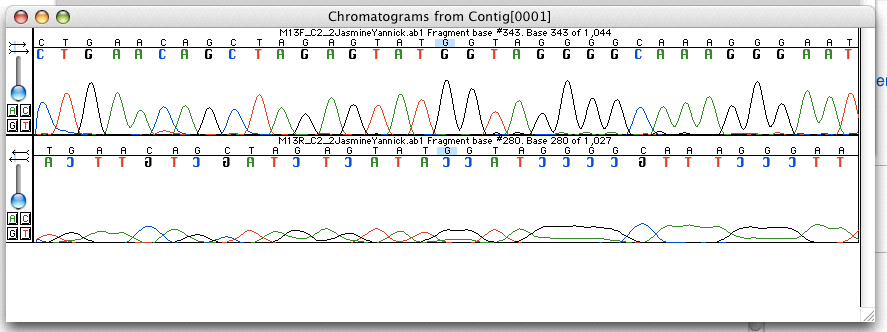
Cancer Genome Sequencing Workflow
Cancer is a group of diseases involving abnormal cell growth with the potential to invade or spread to other parts of the body. These contrast with benign tumors, which do not spread. Possible signs and symptoms include a lump, abnormal bleeding, prolonged cough, unexplained weight loss, and a change in bowel movements. While these symptoms may indicate cancer, they can also have other causes. Over 100 types of cancers affect humans. Tobacco use is the cause of about 22% of cancer deaths. Another 10% are due to obesity, poor diet, lack of physical activity or excessive drinking of alcohol. Other factors include certain infections, exposure to ionizing radiation, and environmental pollutants. In the developing world, 15% of cancers are due to infections such as ''Helicobacter pylori'', hepatitis B, hepatitis C, human papillomavirus infection, Epstein–Barr virus and human immunodeficiency virus (HIV). These factors act, at least partly, by changing the genes of a cell. Ty ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ion Semiconductor Sequencing
Ion semiconductor sequencing is a method of DNA sequencing based on the detection of hydrogen ions that are released during the polymerization of DNA. This is a method of "sequencing by synthesis", during which a complementary strand is built based on the sequence of a template strand. A microwell containing a template DNA strand to be sequenced is flooded with a single species of deoxyribonucleotide triphosphate (dNTP). If the introduced dNTP is complementary to the leading template nucleotide, it is incorporated into the growing complementary strand. This causes the release of a hydrogen ion that triggers an ISFET ion sensor, which indicates that a reaction has occurred. If homopolymer repeats are present in the template sequence, multiple dNTP molecules will be incorporated in a single cycle. This leads to a corresponding number of released hydrogens and a proportionally higher electronic signal. This technology differs from other sequencing-by-synthesis technologies in that n ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Nanopore Sequencing
Nanopore sequencing is a third generation approach used in the sequencing of biopolymers — specifically, polynucleotides in the form of DNA or RNA. Using nanopore sequencing, a single molecule of DNA or RNA can be sequenced without the need for PCR amplification or chemical labeling of the sample. Nanopore sequencing has the potential to offer relatively low-cost genotyping, high mobility for testing, and rapid processing of samples with the ability to display results in real-time. Publications on the method outline its use in rapid identification of viral pathogens, monitoring ebola, environmental monitoring, food safety monitoring, human genome sequencing, plant genome sequencing, monitoring of antibiotic resistance, haplotyping and other applications. Development Nanopore sequencing took 25 years to fully materialize. It involved close collaboration between academia and industry. One of the first people to put forward the idea for nanopore sequencing was David D ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Single Molecule Real Time Sequencing
Single-molecule real-time (SMRT) sequencing is a parallelized single molecule DNA sequencing method. Single-molecule real-time sequencing utilizes a zero-mode waveguide (ZMW). A single DNA polymerase enzyme is affixed at the bottom of a ZMW with a single molecule of DNA as a template. The ZMW is a structure that creates an illuminated observation volume that is small enough to observe only a single nucleotide of DNA being incorporated by DNA polymerase. Each of the four DNA bases is attached to one of four different fluorescent dyes. When a nucleotide is incorporated by the DNA polymerase, the fluorescent tag is cleaved off and diffuses out of the observation area of the ZMW where its fluorescence is no longer observable. A detector detects the fluorescent signal of the nucleotide incorporation, and the base call is made according to the corresponding fluorescence of the dye. Technology The DNA sequencing is done on a chip that contains many ZMWs. Inside each ZMW, a single active ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Illumina (company)
Illumina, Inc. is an American biotechnology company, headquartered in San Diego, California. Incorporated on April 1, 1998, Illumina develops, manufactures, and markets integrated systems for the analysis of genetic variation and biological function. The company provides a line of products and services that serves the sequencing, genotyping and gene expression, and proteomics markets. Illumina's technology had purportedly reduced the cost of sequencing a human genome to by 2014. Its customers include genomic research centers, pharmaceutical companies, academic institutions, clinical research organizations, and biotechnology companies. History Illumina was founded in April 1998 by David Walt, Larry Bock, John Stuelpnagel, Anthony Czarnik, and Mark Chee. While working with CW Group, a venture-capital firm, Bock and Stuelpnagel uncovered what would become Illumina's BeadArray technology at Tufts University and negotiated an exclusive license to that technology. In 1999, Illumin ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
ABI Solid Sequencing
SOLiD (Sequencing by Oligonucleotide Ligation and Detection) is a next-generation DNA sequencing technology developed by Life Technologies and has been commercially available since 2006. This next generation technology generates 108 - 109 small sequence reads at one time. It uses 2 base encoding to decode the raw data generated by the sequencing platform into sequence data. This method should not be confused with "sequencing by synthesis," a principle used by Roche-454 pyrosequencing (introduced in 2005, generating millions of 200-400bp reads in 2009), and the Solexa system (now owned by Illumina) (introduced in 2006, generating hundreds of millions of 50-100bp reads in 2009) These methods have reduced the cost from $0.01/base in 2004 to nearly $0.0001/base in 2006 and increased the sequencing capacity from 1,000,000 bases/machine/day in 2004 to more than 5,000,000,000 bases/machine/day in 2009. Over 30 publications exist describing its use first for nucleosome positioning ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Pyrosequencing
Pyrosequencing is a method of DNA sequencing (determining the order of nucleotides in DNA) based on the "sequencing by synthesis" principle, in which the sequencing is performed by detecting the nucleotide incorporated by a DNA polymerase. Pyrosequencing relies on light detection based on a chain reaction when pyrophosphate is released. Hence, the name pyrosequencing. The principle of pyrosequencing was first described in 1993 by, Bertil Pettersson, Mathias Uhlen and Pål Nyren by combining the solid phase sequencing method using streptavidin coated magnetic beads with recombinant DNA polymerase lacking 3´to 5´exonuclease activity (proof-reading) and luminescence detection using the firefly luciferase enzyme. A mixture of three enzymes (DNA polymerase, ATP sulfurylase and firefly luciferase) and a nucleotide (dNTP) are added to single stranded DNA to be sequenced and the incorporation of nucleotide is followed by measuring the light emitted. The intensity of the light determi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
3rd Generation Sequencing
Third or 3rd may refer to: Numbers * 3rd, the ordinal form of the cardinal number 3 * , a fraction of one third * 1⁄60 of a ''second'', or 1⁄3600 of a ''minute'' Places * 3rd Street (other) * Third Avenue (other) * Highway 3 Music Music theory *Interval number of three in a musical interval ** major third, a third spanning four semitones ** minor third, a third encompassing three half steps, or semitones **neutral third, wider than a minor third but narrower than a major third **augmented third, an interval of five semitones ** diminished third, produced by narrowing a minor third by a chromatic semitone *Third (chord), chord member a third above the root * Degree (music), three away from tonic **mediant, third degree of the diatonic scale ** submediant, sixth degree of the diatonic scale – three steps below the tonic **chromatic mediant, chromatic relationship by thirds *Ladder of thirds, similar to the circle of fifths Albums *'' Third/Sister Lovers ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |



.jpg)
.jpg)