|
Anti-sigma Factors
In the regulation of gene expression in prokaryotes, anti-sigma factors bind to sigma factors and inhibit transcriptional activity. Anti-sigma factors have been found in a number of bacteria, including ''Escherichia coli'' and ''Salmonella'', and in the T4 bacteriophage. Anti-sigma factors are antagonists to the sigma factors, which regulate numerous cell processes including flagellar production, stress response, transport and cellular growth. For example, anti-sigma factor 70 Rsd in ''E. coli'' is present in the stationary phase and blocks the activity of sigma factor 70 which in essence initiates gene transcription. This allows the sigma S factor to associate with RNA polymerase and direct the expression of the stationary genes. Although binding of Rsd to σ70 has been shown and numerous structural studies on Rsd have been performed, the detailed mechanism of action is still unknown. General information Sigma factor is an important protein which starts the transcription by bi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Audrey Stevens' Inhibtor
Audrey () is an English feminine given name. It is the Anglo-Norman form of the Anglo-Saxon name ''Æðelþryð'', composed of the elements '' æðel'' "noble" and ''þryð'' "strength". The Anglo-Norman form of the name was applied to Saint Audrey (d. 679), also known by the historical form of her name as Saint Æthelthryth. The same name also survived into the modern period in its Anglo-Saxon form, as ''Etheldred'', e.g. Etheldred Benett (1776–1845). In the 17th century, the name of ''Saint Audrey'' gave rise to the adjective ''tawdry'' "cheap and pretentious; cheaply adorned". The lace necklaces sold to pilgrims to Saint Audrey fell out of fashion in the 17th century, and so tawdry was reinterpreted as meaning cheap or vulgar. As a consequence, use of the name declined, but it was revived in the 19th century. Popularity of the name in the United States peaked in the interbellum period, but it fell below rank 100 in popularity by 1940 and was not frequently given in the later ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Sigma Factor
A sigma factor (σ factor or specificity factor) is a protein needed for initiation of transcription in bacteria. It is a bacterial transcription initiation factor that enables specific binding of RNA polymerase (RNAP) to gene promoters. It is homologous to archaeal transcription factor B and to eukaryotic factor TFIIB. The specific sigma factor used to initiate transcription of a given gene will vary, depending on the gene and on the environmental signals needed to initiate transcription of that gene. Selection of promoters by RNA polymerase is dependent on the sigma factor that associates with it. They are also found in plant chloroplasts as a part of the bacteria-like plastid-encoded polymerase (PEP). The sigma factor, together with RNA polymerase, is known as the RNA polymerase holoenzyme. Every molecule of RNA polymerase holoenzyme contains exactly one sigma factor subunit, which in the model bacterium ''Escherichia coli'' is one of those listed below. The number of sigma f ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
RpoN
The gene ''rpoN'' (RNA polymerase, nitrogen-limitation N) encodes the sigma factor ''sigma-54'' (σ54, sigma N, or RpoN), a protein in ''Escherichia coli ''Escherichia coli'' (),Wells, J. C. (2000) Longman Pronunciation Dictionary. Harlow ngland Pearson Education Ltd. also known as ''E. coli'' (), is a Gram-negative, facultative anaerobic, rod-shaped, coliform bacterium of the genus ''Escher ...'' and other species of bacteria. RpoN antagonizes RpoS sigma factors. Biological role Originally identified as a regulator of genes involved in nitrogen metabolism and assimilation under nitrogen limiting conditions, ''E. coli'' σ54 has since been shown to play important regulatory roles in a variety of other cellular processes. Similarly, σ54 homologues in other species regulate a wide range of processes, including flagellar synthesis and virulence. Sequence specificity and mechanism of action σ54 promoter elements consist of conserved nucleotides located at −12 and −2 ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
RpoS
The gene ''rpoS'' (RNA polymerase, sigma S, also called katF) encodes the sigma factor ''sigma-38'' (σ38, or RpoS), a 37.8 kD protein in ''Escherichia coli''. Sigma factors are proteins that regulate transcription in bacteria. Sigma factors can be activated in response to different environmental conditions. ''rpoS'' is transcribed in late exponential phase, and RpoS is the primary regulator of stationary phase genes. RpoS is a central regulator of the general stress response and operates in both a retroactive and a proactive manner: it not only allows the cell to survive environmental challenges, but it also prepares the cell for subsequent stresses (cross-protection). The transcriptional regulator CsgD is central to biofilm formation, controlling the expression of the curli structural and export proteins, and the diguanylate cyclase, adrA, which indirectly activates cellulose production. The ''rpoS'' gene most likely originated in the gammaproteobacteria. Environmental signal ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
RpoF
The gene ''rpoF'' (RNA polymerase, flagellum F) encodes the sigma factor ''sigma-28'' (σ28, or RpoF), a protein in ''Escherichia coli'' and other species of bacteria. Depending on the bacterial species, this gene may be referred to as ''sigD'' or ''fliA''. The protein encoded by this gene has been found to be necessary for flagellum A flagellum (; ) is a hairlike appendage that protrudes from certain plant and animal sperm cells, and from a wide range of microorganisms to provide motility. Many protists with flagella are termed as flagellates. A microorganism may have f ... formation. References Escherichia coli genes {{gene-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
RpoE
The gene ''rpoE'' (RNA polymerase, extracytoplasmic E) encodes the sigma factor ''sigma-24'' (σ24, sigma E, or RpoE), a protein in ''Escherichia coli'' and other species of bacteria. Depending on the bacterial species, this gene may be referred to as ''sigE''. RpoE appears to be necessary for the exocytoplasmic stress response. ''E. coli'' mutants without ''rpoE'' cannot grow at high temperatures (that is, above 42 degrees C) and show growth defects at lower temperatures, though this may be due to compensatory mutations. In some bacterial species, such as ''Clostridium botulinum'', this sigma factor may be necessary for sporulation. The stress response regulation activities of RpoE are modulated by the Hfq protein The Hfq protein (also known as HF-I protein) encoded by the ''hfq'' gene was discovered in 1968 as an ''Escherichia coli'' host factor that was essential for replication of the bacteriophage Qβ. It is now clear that Hfq is an abundant bacterial R ... in ''E. coli''. ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Negative Feedback
Negative feedback (or balancing feedback) occurs when some function (Mathematics), function of the output of a system, process, or mechanism is feedback, fed back in a manner that tends to reduce the fluctuations in the output, whether caused by changes in the input or by other disturbances. Whereas positive feedback tends to lead to instability via exponential growth, oscillation or chaos theory, chaotic behavior, negative feedback generally promotes stability. Negative feedback tends to promote a settling to List of types of equilibrium, equilibrium, and reduces the effects of perturbations. Negative feedback loops in which just the right amount of correction is applied with optimum timing can be very stable, accurate, and responsive. Negative feedback is widely used in mechanical and electronic engineering, and also within living organisms, and can be seen in many other fields from chemistry and economics to physical systems such as the climate. General negative feedback ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
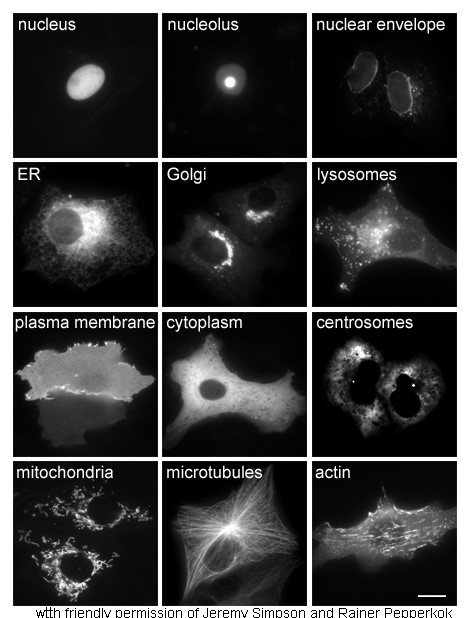
Cytoplasm
In cell biology, the cytoplasm is all of the material within a eukaryotic cell, enclosed by the cell membrane, except for the cell nucleus. The material inside the nucleus and contained within the nuclear membrane is termed the nucleoplasm. The main components of the cytoplasm are cytosol (a gel-like substance), the organelles (the cell's internal sub-structures), and various cytoplasmic inclusions. The cytoplasm is about 80% water and is usually colorless. The submicroscopic ground cell substance or cytoplasmic matrix which remains after exclusion of the cell organelles and particles is groundplasm. It is the hyaloplasm of light microscopy, a highly complex, polyphasic system in which all resolvable cytoplasmic elements are suspended, including the larger organelles such as the ribosomes, mitochondria, the plant plastids, lipid droplets, and vacuoles. Most cellular activities take place within the cytoplasm, such as many metabolic pathways including glycolysis, and proces ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Anti-sigma Factor
In the regulation of gene expression in prokaryotes, anti-sigma factors bind to sigma factors and inhibit transcriptional activity. Anti-sigma factors have been found in a number of bacteria, including ''Escherichia coli'' and ''Salmonella'', and in the T4 bacteriophage. Anti-sigma factors are antagonists to the sigma factors, which regulate numerous cell processes including flagellar production, stress response, transport and cellular growth. For example, anti-sigma factor 70 Rsd in ''E. coli'' is present in the stationary phase and blocks the activity of sigma factor 70 which in essence initiates gene transcription. This allows the sigma S factor to associate with RNA polymerase and direct the expression of the stationary genes. Although binding of Rsd to σ70 has been shown and numerous structural studies on Rsd have been performed, the detailed mechanism of action is still unknown. General information Sigma factor is an important protein which starts the transcription by bi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Gene Expression
Gene expression is the process by which information from a gene is used in the synthesis of a functional gene product that enables it to produce end products, protein or non-coding RNA, and ultimately affect a phenotype, as the final effect. These products are often proteins, but in non-protein-coding genes such as transfer RNA (tRNA) and small nuclear RNA (snRNA), the product is a functional non-coding RNA. Gene expression is summarized in the central dogma of molecular biology first formulated by Francis Crick in 1958, further developed in his 1970 article, and expanded by the subsequent discoveries of reverse transcription and RNA replication. The process of gene expression is used by all known life—eukaryotes (including multicellular organisms), prokaryotes (bacteria and archaea), and utilized by viruses—to generate the macromolecular machinery for life. In genetics, gene expression is the most fundamental level at which the genotype gives rise to the phenotype, '' ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
RNA Polymerase
In molecular biology, RNA polymerase (abbreviated RNAP or RNApol), or more specifically DNA-directed/dependent RNA polymerase (DdRP), is an enzyme that synthesizes RNA from a DNA template. Using the enzyme helicase, RNAP locally opens the double-stranded DNA so that one strand of the exposed nucleotides can be used as a template for the synthesis of RNA, a process called transcription. A transcription factor and its associated transcription mediator complex must be attached to a DNA binding site called a promoter region before RNAP can initiate the DNA unwinding at that position. RNAP not only initiates RNA transcription, it also guides the nucleotides into position, facilitates attachment and elongation, has intrinsic proofreading and replacement capabilities, and termination recognition capability. In eukaryotes, RNAP can build chains as long as 2.4 million nucleotides. RNAP produces RNA that, functionally, is either for protein coding, i.e. messenger RNA (mRNA); or n ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
T4 Bacteriophage
T4 or T-4 may refer to: Airports and airlines * Heathrow Terminal 4 * Tiyas Military Airbase, also known as the T-4 Airbase Biology and medicine * T4 phage, a bacteriophage * Thyroxine (T4), a form of thyroid hormone * the T4 spinal nerve * the fourth thoracic vertebrae of the vertebral column * A non-small cell lung carcinoma staging for a type of tumour * A CD4 + T lymphocyte * T4: an EEG electrode site according to the 10-20 system Entertainment * ''T4'' (Channel 4), the former daytime teen-aimed slot on Channel 4 in the UK * ''Terminator Salvation'', sometimes referred to as ''Terminator 4'' * '' Transformers: Age of Extinction'', the fourth film in the live-action ''Transformers'' film series Computing * SPARC T4, a microprocessor introduced by Oracle Microelectronics in 2011 Software and video games * Text Template Transformation Toolkit, a technology developed by Microsoft * ''Tekken 4'', a fighting game Electricity production * Lockheed Martin's High beta ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |