|
Vps34
Class III PI 3-kinase is a subgroup of the enzyme family, phosphoinositide 3-kinase that share a common protein domain structure, substrate specificity and method of activation. There is only one known class III PI 3-kinase, Vps34, which is also the only PI 3-kinase expressed in all eukaryotic cells. In humans it is encoded by the PIK3C3 gene. In human cells Vps34 associates with a regulatory subunit, PIK3R4(p150, Vps15). Substrate specificity Vps34 is more accurately described as a phosphatidylinositol 3-kinase. ''In vivo'' Vps34 can phosphorylate only phosphatidylinositol to form phosphatidylinositol (3)-phosphate ( PtdIns(3)P). Functions Vps34 was first identified in a ''Saccharomyces cerevisiae'' (budding yeast) screen for proteins involved vesicle-mediated vacuolar protein sorting (hence Vps). A number of proteins containing a phosphoinositide binding domain specific for PtdIns(3)P that function in cellular protein trafficking have been identified. Vps34 has been shown to i ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Phosphoinositide 3-kinase
Phosphoinositide 3-kinases (PI3Ks), also called phosphatidylinositol 3-kinases, are a family of enzymes involved in cellular functions such as cell growth, proliferation, differentiation, motility, survival and intracellular trafficking, which in turn are involved in cancer. PI3Ks are a family of related intracellular signal transducer enzymes capable of phosphorylating the 3 position hydroxyl group of the inositol ring of phosphatidylinositol (PtdIns). The pathway, with oncogene PIK3CA and tumor suppressor gene PTEN, is implicated in the sensitivity of cancer tumors to insulin and IGF1, and in calorie restriction. Discovery The discovery of PI3Ks by Lewis Cantley and colleagues began with their identification of a previously unknown phosphoinositide kinase associated with the polyoma middle T protein. They observed unique substrate specificity and chromatographic properties of the products of the lipid kinase, leading to the discovery that this phosphoinositide kinase had ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
PIK3C3
Phosphatidylinositol 3-kinase catalytic subunit type 3 is an enzyme subunit that in humans is encoded by the ''PIK3C3'' gene In biology, the word gene (from , ; "... Wilhelm Johannsen coined the word gene to describe the Mendelian units of heredity..." meaning ''generation'' or ''birth'' or ''gender'') can have several different meanings. The Mendelian gene is a b .... It's a class III phosphoinositide 3-kinase. References Further reading * * * * * * * * * * * * * * * * * * {{gene-18-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
PIK3R4
Phosphoinositide 3-kinase regulatory subunit 4, also known as PI3-kinase regulatory subunit 4 or PI3-kinase p150 subunit or phosphoinositide 3-kinase adaptor protein, or ''VPS15'' is an enzyme that in humans is encoded by the ''PIK3R4'' gene In biology, the word gene (from , ; "... Wilhelm Johannsen coined the word gene to describe the Mendelian units of heredity..." meaning ''generation'' or ''birth'' or ''gender'') can have several different meanings. The Mendelian gene is a b .... References Further reading * * * * * * * * * * EC 2.7.11 {{gene-3-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
PtdIns(3)P
Phosphatidylinositol 3-phosphate (PtdIns3''P'') is a phospholipid found in cell membranes that helps to recruit a range of proteins, many of which are involved in protein trafficking, to the membranes. It is the product of both the class II and III phosphoinositide 3-kinases (PI 3-kinases) activity on phosphatidylinositol. PtdIns3''P'' is dephosphorylated by the myotubularin family of phosphatases, on the D3 position of the inositol ring, and can be converted to PtdIns(3,5)''P''2 by the lipid kinase PIKfyve. Both FYVE domains and PX domains – found in proteins such as SNX1, HGS, and EEA1 – bind to PtdIns3''P''. The majority of PtdIns3''P'' appears to be constitutively synthesised by the class III PI 3-kinase, PIK3C3 (Vps34), at endocytic membranes. Class II PI 3-kinases also appear to synthesise PtdIns3''P'', their activity however appears to be regulated by a range of stimuli, including growth factors. This suggests that specific pools of PtdIns3''P'' may be ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
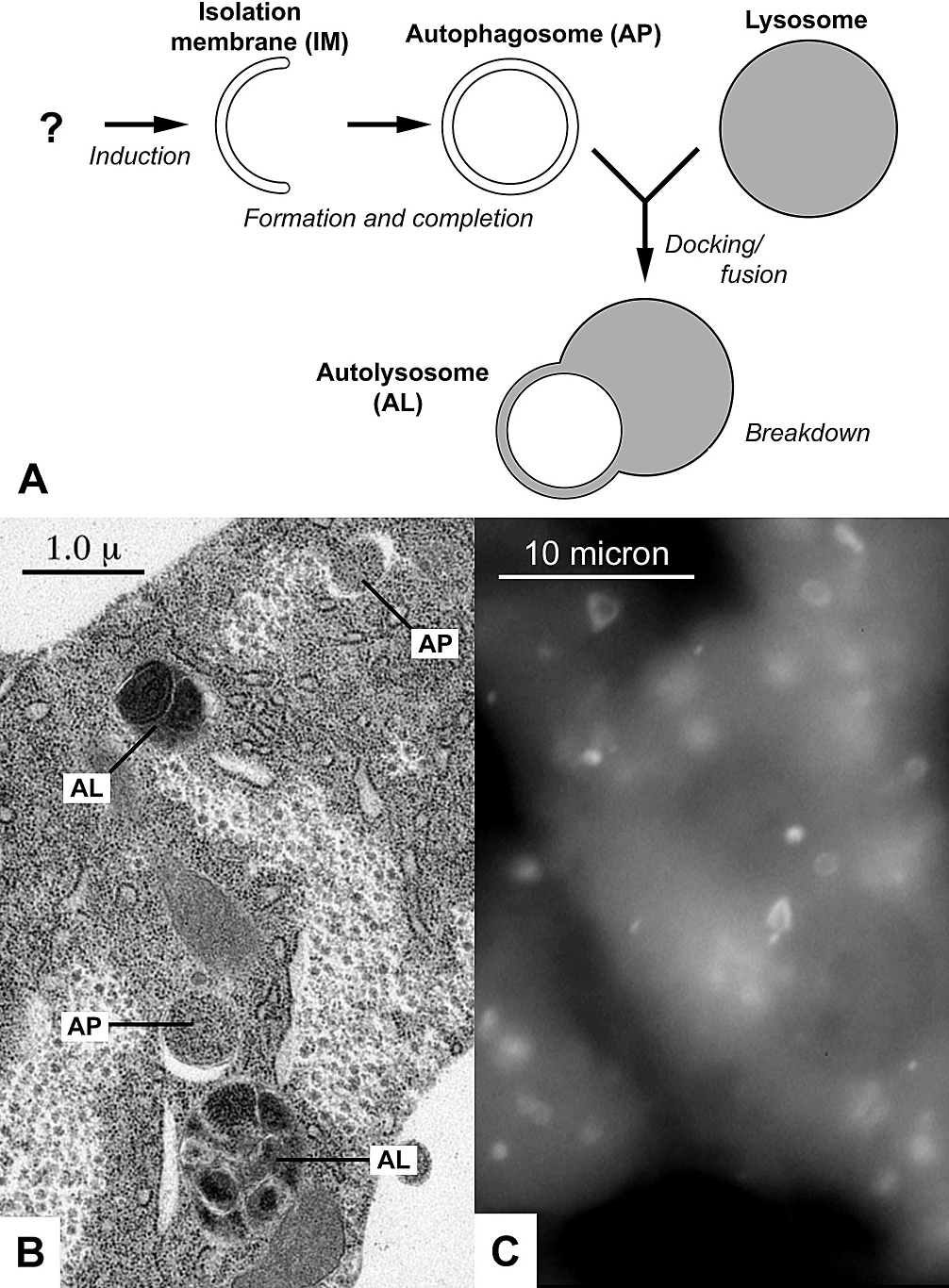
Autophagy
Autophagy (or autophagocytosis; from the Ancient Greek , , meaning "self-devouring" and , , meaning "hollow") is the natural, conserved degradation of the cell that removes unnecessary or dysfunctional components through a lysosome-dependent regulated mechanism. It allows the orderly degradation and recycling of cellular components. Although initially characterized as a primordial degradation pathway induced to protect against starvation, it has become increasingly clear that autophagy also plays a major role in the homeostasis of non-starved cells. Defects in autophagy have been linked to various human diseases, including neurodegeneration and cancer, and interest in modulating autophagy as a potential treatment for these diseases has grown rapidly. Four forms of autophagy have been identified: macroautophagy, microautophagy, chaperone-mediated autophagy (CMA), and crinophagy. In macroautophagy (the most thoroughly researched form of autophagy), cytoplasmic components (like ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Calmodulin
Calmodulin (CaM) (an abbreviation for calcium-modulated protein) is a multifunctional intermediate calcium-binding messenger protein expressed in all eukaryotic cells. It is an intracellular target of the secondary messenger Ca2+, and the binding of Ca2+ is required for the activation of calmodulin. Once bound to Ca2+, calmodulin acts as part of a calcium signal transduction pathway by modifying its interactions with various target proteins such as kinases or phosphatases. Structure Calmodulin is a small, highly conserved protein that is 148 amino acids long (16.7 kDa). The protein has two approximately symmetrical globular domains (the N- and C- domains) each containing a pair of EF hand motifs separated by a flexible linker region for a total of four Ca2+ binding sites, two in each globular domain. In the Ca2+-free state, the helices that form the four EF-hands are collapsed in a compact orientation, and the central linker is disordered; in the Ca2+-saturated state, ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
EGTA (chemical)
EGTA (ethylene glycol-bis(β-aminoethyl ether)-''N'',''N'',''N''′,''N''′-tetraacetic acid), also known as egtazic acid (INN, USAN), is an aminopolycarboxylic acid, a chelating agent. It is a white solid that is related to the better known EDTA. Compared to EDTA, it has a lower affinity for magnesium, making it more selective for calcium ions. It is useful in buffer solutions that resemble the environment in living cells where calcium ions are usually at least a thousandfold less concentrated than magnesium. The p''K''a for binding of calcium ions by tetrabasic EGTA is 11.00, but the protonated forms do not significantly contribute to binding, so at pH 7, the apparent p''K''a becomes 6.91. See Qin ''et al.'' for an example of a p''K''a calculation. EGTA has also been used experimentally for the treatment of animals with cerium poisoning and for the separation of thorium from the mineral monazite. EGTA is used as a compound in elution buffer in the protein purification ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Drosophila
''Drosophila'' () is a genus of flies, belonging to the family Drosophilidae, whose members are often called "small fruit flies" or (less frequently) pomace flies, vinegar flies, or wine flies, a reference to the characteristic of many species to linger around overripe or rotting fruit. They should not be confused with the Tephritidae, a related family, which are also called fruit flies (sometimes referred to as "true fruit flies"); tephritids feed primarily on unripe or ripe fruit, with many species being regarded as destructive agricultural pests, especially the Mediterranean fruit fly. One species of ''Drosophila'' in particular, '' D. melanogaster'', has been heavily used in research in genetics and is a common model organism in developmental biology. The terms "fruit fly" and "''Drosophila''" are often used synonymously with ''D. melanogaster'' in modern biological literature. The entire genus, however, contains more than 1,500 species and is very diverse in appea ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
FYVE Domain
In molecular biology the FYVE zinc finger domain is named after the four cysteine-rich proteins: Fab 1 (yeast orthologue of PIKfyve), YOTB, Vac 1 (vesicle transport protein), and EEA1, in which it has been found. FYVE domains bind phosphatidylinositol 3-phosphate, in a way dependent on its metal ion coordination and basic amino acids. The FYVE domain inserts into cell membranes in a pH-dependent manner. The FYVE domain has been connected to vacuolar protein sorting and endosome function. Structure The FYVE domain is composed of two small beta hairpins (or zinc knuckles) followed by an alpha helix. The FYVE finger binds two zinc ions. The FYVE finger has eight potential zinc coordinating cysteine positions and is characterized by having basic amino acids around the cysteines. Many members of this family also include two histidines in a sequence motif: The FYVE finger is structurally similar to the RING domain and the PHD finger. Examples The following is a list of human p ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
SiRNA
Small interfering RNA (siRNA), sometimes known as short interfering RNA or silencing RNA, is a class of double-stranded RNA at first non-coding RNA molecules, typically 20-24 (normally 21) base pairs in length, similar to miRNA, and operating within the RNA interference (RNAi) pathway. It interferes with the expression of specific genes with complementary nucleotide sequences by degrading mRNA after transcription, preventing translation. Text was copied from this source, which is available under Creative Commons Attribution 4.0 International License Structure Naturally occurring siRNAs have a well-defined structure that is a short (usually 20 to 24- bp) double-stranded RNA (dsRNA) with phosphorylated 5' ends and hydroxylated 3' ends with two overhanging nucleotides. The Dicer enzyme catalyzes production of siRNAs from long dsRNAs and small hairpin RNAs. siRNAs can also be introduced into cells by transfection. Since in principle any gene can be knocked down by a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
MTORC1
mTORC1, also known as mammalian target of rapamycin complex 1 or mechanistic target of rapamycin complex 1, is a protein complex that functions as a nutrient/energy/redox sensor and controls protein synthesis. mTOR Complex 1 (mTORC1) is composed of the mTOR protein complex, regulatory-associated protein of mTOR (commonly known as raptor), mammalian lethal with SEC13 protein 8 (MLST8), PRAS40 and DEPTOR. This complex embodies the classic functions of mTOR, namely as a nutrient/energy/redox sensor and controller of protein synthesis. The activity of this complex is regulated by rapamycin, insulin, growth factors, phosphatidic acid, certain amino acids and their derivatives (e.g., -leucine and β-hydroxy β-methylbutyric acid), mechanical stimuli, and oxidative stress. Recently it has been also demonstrated that cellular bicarbonate metabolism can be regulated by mTORC1 signaling. The role of mTORC1 is to activate translation of proteins. In order for cells to grow and prolife ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
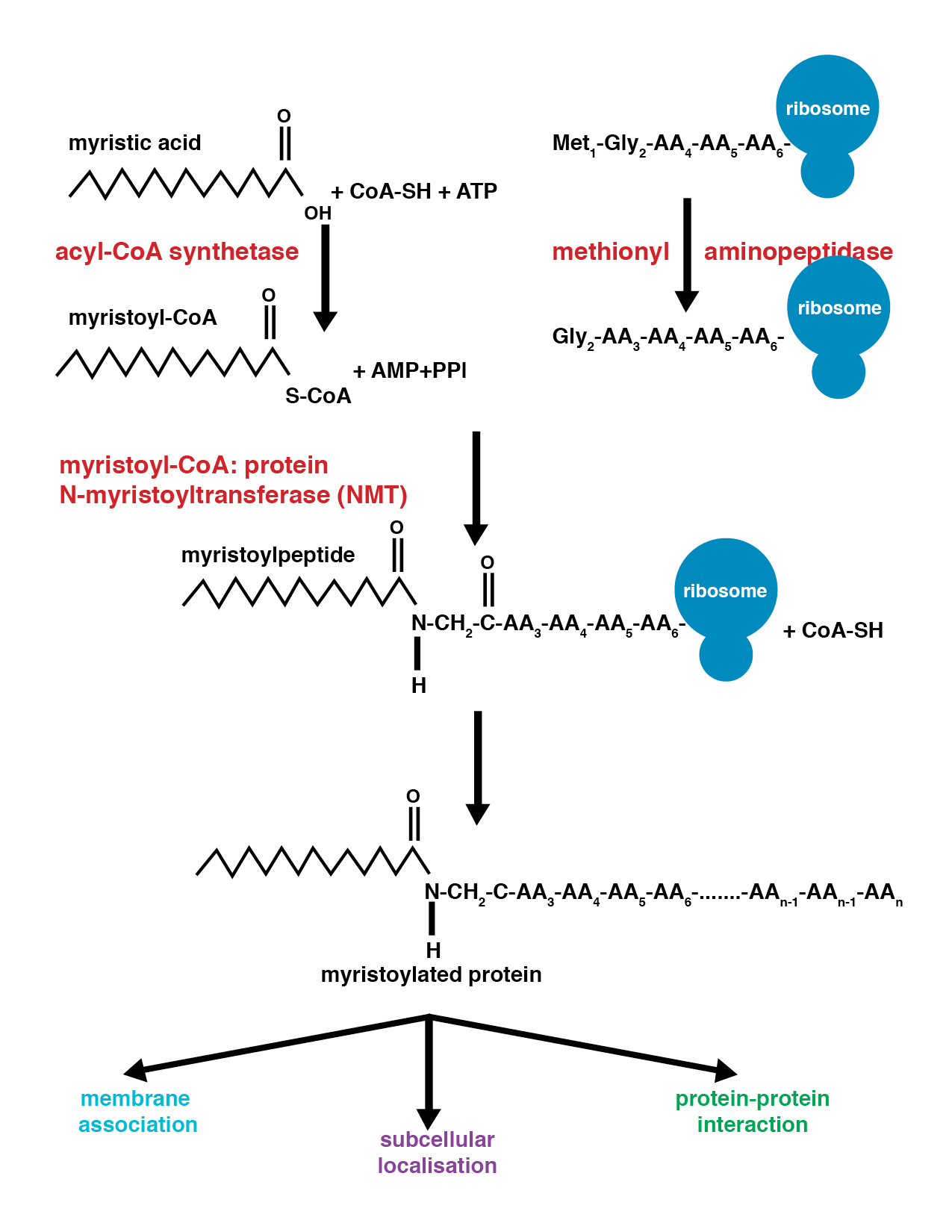
Myristoylation
Myristoylation is a lipidation modification where a myristoyl group, derived from myristic acid, is covalently attached by an amide bond to the alpha-amino group of an N-terminal glycine residue. Myristic acid is a 14-carbon saturated fatty acid (14:0) with the systematic name of ''n''-Tetradecanoic acid. This modification can be added either co-translationally or post-translationally. N-myristoyltransferase (NMT) catalyzes the myristic acid addition reaction in the cytoplasm of cells. This lipidation event is the most found type of fatty acylation and is common among many organisms including animals, plants, fungi, protozoans and viruses. Myristoylation allows for weak protein–protein and protein–lipid interactions and plays an essential role in membrane targeting, protein–protein interactions and functions widely in a variety of signal transduction pathways. Discovery In 1982, Koiti Titani's lab identified an "N-terminal blocking group" on the catalytic subunit of ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |