|
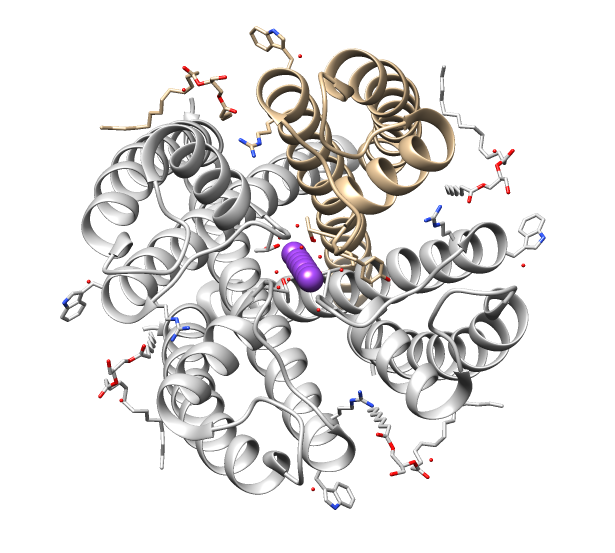
VDAC
Voltage-dependent anion channels, or mitochondrial porins, are a class of porin ion channel located on the outer mitochondrial membrane. There is debate as to whether or not this channel is expressed in the cell surface membrane. This major protein of the outer mitochondrial membrane of eukaryotes forms a voltage-dependent anion-selective channel (VDAC) that behaves as a general diffusion pore for small hydrophilic molecules. The channel adopts an open conformation at low or zero membrane potential and a closed conformation at potentials above 30–40 mV. VDAC facilitates the exchange of ions and molecules between mitochondria and cytosol and is regulated by the interactions with other proteins and small molecules. Structure This protein contains about 280 amino acids and forms a beta barrel which spans the mitochondrial outer membrane. Since its discovery in 1976, extensive function and structure analysis of VDAC proteins has been conducted. A prominent feature of the ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Outer Mitochondrial Membrane
A mitochondrion (; ) is an organelle found in the cells of most Eukaryotes, such as animals, plants and fungi. Mitochondria have a double membrane structure and use aerobic respiration to generate adenosine triphosphate (ATP), which is used throughout the cell as a source of chemical energy. They were discovered by Albert von Kölliker in 1857 in the voluntary muscles of insects. The term ''mitochondrion'' was coined by Carl Benda in 1898. The mitochondrion is popularly nicknamed the "powerhouse of the cell", a phrase coined by Philip Siekevitz in a 1957 article of the same name. Some cells in some multicellular organisms lack mitochondria (for example, mature mammalian red blood cells). A large number of unicellular organisms, such as microsporidia, parabasalids and diplomonads, have reduced or transformed their mitochondria into other structures. One eukaryote, ''Monocercomonoides'', is known to have completely lost its mitochondria, and one multicellular organism, '' Henn ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Mitochondria
A mitochondrion (; ) is an organelle found in the Cell (biology), cells of most Eukaryotes, such as animals, plants and Fungus, fungi. Mitochondria have a double lipid bilayer, membrane structure and use aerobic respiration to generate adenosine triphosphate (ATP), which is used throughout the cell as a source of chemical energy. They were discovered by Albert von Kölliker in 1857 in the voluntary muscles of insects. The term ''mitochondrion'' was coined by Carl Benda in 1898. The mitochondrion is popularly nicknamed the "powerhouse of the cell", a phrase coined by Philip Siekevitz in a 1957 article of the same name. Some cells in some multicellular organisms lack mitochondria (for example, mature mammalian red blood cells). A large number of unicellular organisms, such as microsporidia, parabasalids and diplomonads, have reduced or transformed their mitochondria into mitosome, other structures. One eukaryote, ''Monocercomonoides'', is known to have completely lost its mitocho ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Bcl-2-associated X Protein
Apoptosis regulator BAX, also known as bcl-2-like protein 4, is a protein that in humans is encoded by the ''BAX'' gene. ''BAX'' is a member of the Bcl-2 gene family. BCL2 family members form hetero- or homodimers and act as anti- or pro-apoptotic regulators that are involved in a wide variety of cellular activities. This protein forms a heterodimer with BCL2, and functions as an apoptotic activator. This protein is reported to interact with, and increase the opening of, the mitochondrial voltage-dependent anion channel (VDAC), which leads to the loss in membrane potential and the release of cytochrome c. The expression of this gene is regulated by the tumor suppressor P53 and has been shown to be involved in P53-mediated apoptosis. Structure The ''BAX'' gene was the first identified pro-apoptotic member of the Bcl-2 protein family. Bcl-2 family members share one or more of the four characteristic domains of homology entitled the Bcl-2 homology (BH) domains (named BH1, BH2, B ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Mitochondrial Permeability Transition Pore
The mitochondrial permeability transition pore (mPTP or MPTP; also referred to as PTP, mTP or MTP) is a protein that is formed in the inner membrane of the mitochondria under certain pathological conditions such as traumatic brain injury and stroke. Opening allows increase in the permeability of the mitochondrial membranes to molecules of less than 1500 Daltons in molecular weight. Induction of the permeability transition pore, mitochondrial membrane permeability transition (mPT or MPT), can lead to mitochondrial swelling and cell death through apoptosis or necrosis depending on the particular biological setting. Roles in pathology The MPTP was originally discovered by Haworth and Hunter in 1979 and has been found to be involved in neurodegeneration, hepatotoxicity from Reye-related agents, cardiac necrosis and nervous and muscular dystrophies among other deleterious events inducing cell damage and death. MPT is one of the major causes of cell death in a variety of conditions. ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cytochrome C
The cytochrome complex, or cyt ''c'', is a small hemeprotein found loosely associated with the inner membrane of the mitochondrion. It belongs to the cytochrome c family of proteins and plays a major role in cell apoptosis. Cytochrome c is highly water-soluble, unlike other cytochromes, and is an essential component of the respiratory electron transport chain, where it carries one electron. It is capable of undergoing oxidation and reduction as its iron atom converts between the ferrous and ferric forms, but does not bind oxygen. It transfers electrons between Complexes III (Coenzyme Q – Cyt c reductase) and IV (Cyt c oxidase). In humans, cytochrome c is encoded by the ''CYCS'' gene. Species distribution Cytochrome c is a highly conserved protein across the spectrum of eukaryotic species, found in plants, animals, fungi, and many unicellular organisms. This, along with its small size (molecular weight about 12,000 daltons), makes it useful in studies of cladistics. Cyt ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ion Channel
Ion channels are pore-forming membrane proteins that allow ions to pass through the channel pore. Their functions include establishing a resting membrane potential, shaping action potentials and other electrical signals by gating the flow of ions across the cell membrane, controlling the flow of ions across secretory and epithelial cells, and regulating cell volume. Ion channels are present in the membranes of all cells. Ion channels are one of the two classes of ionophoric proteins, the other being ion transporters. The study of ion channels often involves biophysics, electrophysiology, and pharmacology, while using techniques including voltage clamp, patch clamp, immunohistochemistry, X-ray crystallography, fluoroscopy, and RT-PCR. Their classification as molecules is referred to as channelomics. Basic features There are two distinctive features of ion channels that differentiate them from other types of ion transporter proteins: #The rate of ion transport through the ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Caspases
Caspases (cysteine-aspartic proteases, cysteine aspartases or cysteine-dependent aspartate-directed proteases) are a family of protease enzymes playing essential roles in programmed cell death. They are named caspases due to their specific cysteine protease activity – a cysteine in its active site nucleophilically attacks and cleaves a target protein only after an aspartic acid residue. As of 2009, there are 12 confirmed caspases in humans and 10 in mice, carrying out a variety of cellular functions. The role of these enzymes in programmed cell death was first identified in 1993, with their functions in apoptosis well characterised. This is a form of programmed cell death, occurring widely during development, and throughout life to maintain cell homeostasis. Activation of caspases ensures that the cellular components are degraded in a controlled manner, carrying out cell death with minimal effect on surrounding tissues. Caspases have other identified roles in programmed cell ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Bcl-2
Bcl-2 (B-cell lymphoma 2), encoded in humans by the ''BCL2'' gene, is the founding member of the Bcl-2 family of regulator proteins that regulate cell death (apoptosis), by either inhibiting (anti-apoptotic) or inducing (pro-apoptotic) apoptosis. It was the first apoptosis regulator identified in any organism. Bcl-2 derives its name from ''B-cell lymphoma 2'', as it is the second member of a range of proteins initially described in chromosomal translocations involving chromosomes 14 and 18 in follicular lymphomas. Orthologs (such as ''Bcl2'' in mice) have been identified in numerous mammals for which complete genome data are available. Like BCL3, BCL5, BCL6, BCL7A, BCL9, and BCL10, it has clinical significance in lymphoma. Isoforms The two isoforms of Bcl-2, Isoform 1, and Isoform 2, exhibit a similar fold. However, results in the ability of these isoforms to bind to the BAD and BAK proteins, as well as in the structural topology and electrostatic potential of the binding ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
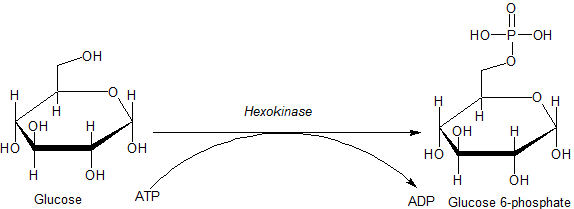
Hexokinase
A hexokinase is an enzyme that phosphorylates hexoses (six-carbon sugars), forming hexose phosphate. In most organisms, glucose is the most important substrate for hexokinases, and glucose-6-phosphate is the most important product. Hexokinase possesses the ability to transfer an inorganic phosphate group from ATP to a substrate. Hexokinases should not be confused with glucokinase, which is a specific isoform of hexokinase. All hexokinases are capable of phosphorylating several hexoses but glucokinase acts with a 50-fold lower substrate affinity and its main hexose substrate is glucose. Variation Genes that encode hexokinase have been discovered in every domain of life, and exist among a variety of species that range from bacteria, yeast, and plants to humans and other vertebrates. They are categorized as ''actin fold'' proteins, sharing a common ATP binding site core that is surrounded by more variable sequences which determine substrate affinities and other properties. ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Isocitrate Dehydrogenase
Isocitrate dehydrogenase (IDH) () and () is an enzyme that catalyzes the oxidative decarboxylation of isocitrate, producing alpha-ketoglutarate (α-ketoglutarate) and CO2. This is a two-step process, which involves oxidation of isocitrate (a secondary alcohol) to oxalosuccinate (a ketone), followed by the decarboxylation of the carboxyl group beta to the ketone, forming alpha-ketoglutarate. In humans, IDH exists in three isoforms: IDH3 catalyzes the third step of the citric acid cycle while converting NAD+ to NADH in the mitochondria. The isoforms IDH1 and IDH2 catalyze the same reaction outside the context of the citric acid cycle and use NADP+ as a cofactor instead of NAD+. They localize to the cytosol as well as the mitochondrion and peroxisome. Isozymes The following is a list of human isocitrate dehydrogenase isozymes: NADP+ dependent Each NADP+-dependent isozyme functions as a homodimer: See also * Isocitrate/isopropylmalate dehydrogenase family NAD+ depe ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
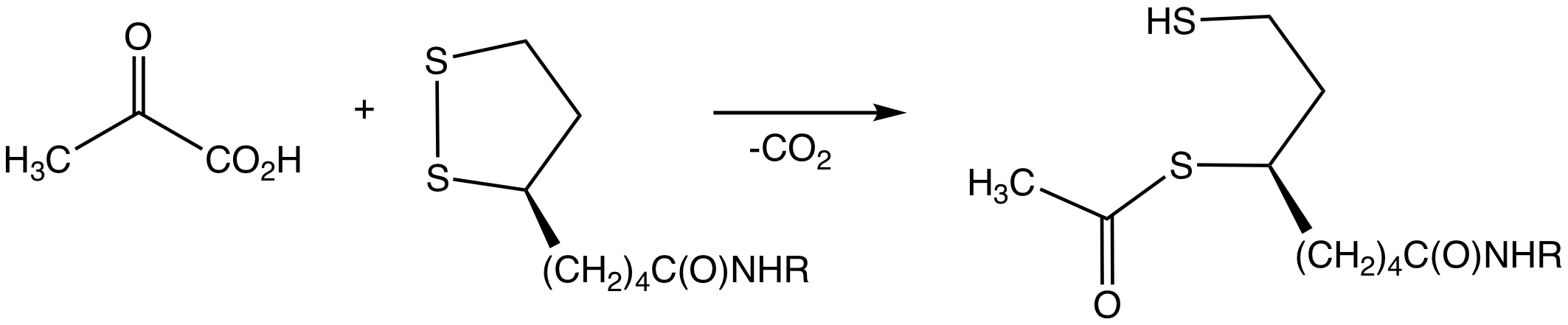
Pyruvate Dehydrogenase
Pyruvate dehydrogenase is an enzyme that catalyzes the reaction of pyruvate and a lipoamide to give the acetylated dihydrolipoamide and carbon dioxide. The conversion requires the coenzyme thiamine pyrophosphate. Pyruvate dehydrogenase is usually encountered as a component, referred to as E1, of the pyruvate dehydrogenase complex (PDC). PDC consists of other enzymes, referred to as E2 and E3. Collectively E1-E3 transform pyruvate, NAD+, coenzyme A into acetyl-CoA, CO2, and NADH. The conversion is crucial because acetyl-CoA may then be used in the citric acid cycle to carry out cellular respiration. To distinguish between this enzyme and the PDC, it is systematically called pyruvate dehydrogenase (acetyl-transferring). Mechanism The thiamine pyrophosphate (TPP) converts to an ylide by deprotonation. The ylide attack the ketone group of pyruvate. The resulting adduct decarboxylates. The resulting 1,3-dipole reductively acetylates lipoamide-E2. In terms of details, bioch ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Oxidative Phosphorylation
Oxidative phosphorylation (UK , US ) or electron transport-linked phosphorylation or terminal oxidation is the metabolic pathway in which cells use enzymes to oxidize nutrients, thereby releasing chemical energy in order to produce adenosine triphosphate (ATP). In eukaryotes, this takes place inside mitochondria. Almost all aerobic organisms carry out oxidative phosphorylation. This pathway is so pervasive because it releases more energy than alternative fermentation processes such as anaerobic glycolysis. The energy stored in the chemical bonds of glucose is released by the cell in the citric acid cycle producing carbon dioxide, and the energetic electron donors NADH and FADH. Oxidative phosphorylation uses these molecules and O2 to produce ATP, which is used throughout the cell whenever energy is needed. During oxidative phosphorylation, electrons are transferred from the electron donors to a series of electron acceptors in a series of redox reactions ending in oxygen, who ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |