|
UCSF Chimera
UCSF Chimera (or simply Chimera) is an extensible program for interactive visualization and analysis of molecular structures and related data, including density maps, supramolecular assemblies, sequence alignments, docking results, trajectories, and conformational ensembles. High-quality images and movies can be created. Chimera includes complete documentation and can be downloaded free of charge for noncommercial use. Chimera is developed by the Resource for Biocomputing, Visualization, and Informatics (RBVI) at the University of California, San Francisco. Development is partially supported by the National Institutes of Health (NIGMS grant P41-GM103311). The next-generation program is UCSF ChimeraX. General structure analysis * automatic identification of atom * hydrogen addition and partial charge assignment * high-quality hydrogen bond, contact, and clash detection * measurements: distances, angles, surface area, volume * calculation of centroids, axes, planes and associat ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Microsoft Windows
Windows is a group of several Proprietary software, proprietary graphical user interface, graphical operating system families developed and marketed by Microsoft. Each family caters to a certain sector of the computing industry. For example, Windows NT for consumers, Windows Server for servers, and Windows IoT for embedded systems. Defunct Windows families include Windows 9x, Windows Mobile, and Windows Phone. The first version of Windows was released on November 20, 1985, as a graphical operating system shell for MS-DOS in response to the growing interest in graphical user interfaces (GUIs). Windows is the most popular desktop operating system in the world, with Usage share of operating systems, 75% market share , according to StatCounter. However, Windows is not the most used operating system when including both mobile and desktop OSes, due to Android (operating system), Android's massive growth. , the most recent version of Windows is Windows 11 for consumer Personal compu ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
UniProt
UniProt is a freely accessible database of protein sequence and functional information, many entries being derived from genome sequencing projects. It contains a large amount of information about the biological function of proteins derived from the research literature. It is maintained by the UniProt consortium, which consists of several European bioinformatics organisations and a foundation from Washington, DC, United States. The UniProt consortium The UniProt consortium comprises the European Bioinformatics Institute (EBI), the Swiss Institute of Bioinformatics (SIB), and the Protein Information Resource (PIR). EBI, located at the Wellcome Trust Genome Campus in Hinxton, UK, hosts a large resource of bioinformatics databases and services. SIB, located in Geneva, Switzerland, maintains the ExPASy (Expert Protein Analysis System) servers that are a central resource for proteomics tools and databases. PIR, hosted by the National Biomedical Research Foundation (NBRF) at the Geo ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Conserved Domain Database
The Conserved Domain Database (CDD) is a database of well-annotated multiple sequence alignment models and derived database search models, for ancient domains and full-length proteins. Philosophy Domains can be thought of as distinct functional and/or structural units of a protein. These two classifications coincide rather often, as a matter of fact, and what is found as an independently folding unit of a polypeptide chain also carries specific function. Domains are often identified as recurring (sequence or structure) units, which may exist in various contexts. In molecular evolution such domains may have been utilized as building blocks, and may have been recombined in different arrangements to modulate protein function. CDD defines conserved domains as recurring units in molecular evolution, the extents of which can be determined by sequence and structure analysis. The goal of the NCBI conserved domain curation project is to provide database users with insights into how patt ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
MODELLER
Modeller, often stylized as MODELLER, is a computer program used for homology modeling to produce models of protein tertiary structures and quaternary structures (rarer). It implements a method inspired by nuclear magnetic resonance spectroscopy of proteins (protein NMR), termed '' satisfaction of spatial restraints'', by which a set of geometrical criteria are used to create a probability density function for the location of each atom in the protein. The method relies on an input sequence alignment between the target amino acid sequence to be modeled and a template protein which structure has been solved. The program also incorporates limited functions for ab initio structure prediction of loop regions of proteins, which are often highly variable even among homologous proteins and thus difficult to predict by homology modeling. Modeller was originally written and is currently maintained by Andrej Sali at the University of California, San Francisco. It runs on the operating s ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
MUSCLE (alignment Software)
MUltiple Sequence Comparison by Log-Expectation (MUSCLE) is computer software for multiple sequence alignment of protein and nucleotide sequences. It is licensed as public domain. The method was published by Robert C. Edgar in two papers in 2004. The first paper, published in ''Nucleic Acids Research'', introduced the sequence alignment algorithm. The second paper, published in ''BMC Bioinformatics'', presented more technical details. Algorithm The MUSCLE algorithm proceeds in three stages: the ''draft progressive'', ''improved progressive'', and ''refinement'' stage. Stage 1: Draft Progressive In this first stage, the algorithm produces a multiple alignment, emphasizing speed over accuracy. This step begins by computing the k-mer distance for every pair of input sequences to create a distance matrix. UPGMA clusters the distance matrix to produce a binary tree. From this tree a progressive alignment is constructed, beginning with the creation of profiles for each leaf of the tre ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Clustal
Clustal is a series of widely used computer programs used in bioinformatics for multiple sequence alignment. There have been many versions of Clustal over the development of the algorithm that are listed below. The analysis of each tool and its algorithm are also detailed in their respective categories. Available operating systems listed in the sidebar are a combination of the software availability and may not be supported for every current version of the Clustal tools. Clustal Omega has the widest variety of operating systems out of all the Clustal tools. History There have been many variations of the Clustal software, all of which are listed below: * Clustal: The original software for multiple sequence alignments, created by Des Higgins in 1988, was based on deriving phylogenetic trees from pairwise sequences of amino acids or nucleotides. * ClustalV: The second generation of the Clustal software was released in 1992 and was a rewrite of the original Clustal package. It int ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
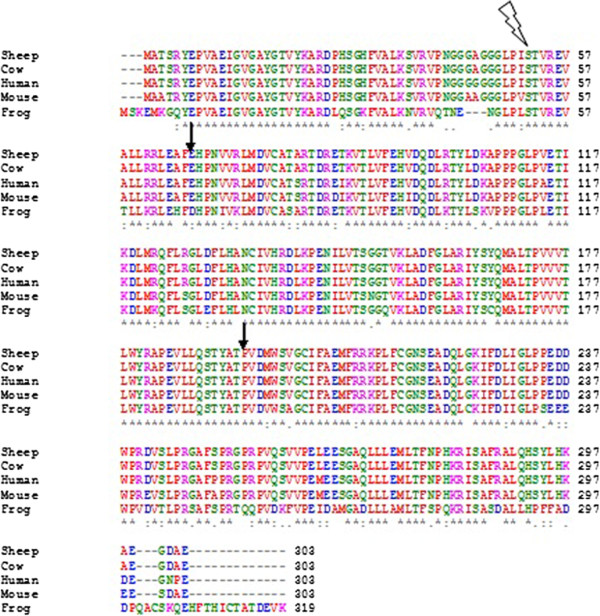
Multiple Sequence Alignment
Multiple sequence alignment (MSA) may refer to the process or the result of sequence alignment of three or more biological sequences, generally protein, DNA, or RNA. In many cases, the input set of query sequences are assumed to have an evolutionary relationship by which they share a linkage and are descended from a common ancestor. From the resulting MSA, sequence homology can be inferred and phylogenetic analysis can be conducted to assess the sequences' shared evolutionary origins. Visual depictions of the alignment as in the image at right illustrate mutation events such as point mutations (single amino acid or nucleotide changes) that appear as differing characters in a single alignment column, and insertion or deletion mutations (indels or gaps) that appear as hyphens in one or more of the sequences in the alignment. Multiple sequence alignment is often used to assess sequence conservation of protein domains, tertiary and secondary structures, and even individual amino aci ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
BLAST
Blast or The Blast may refer to: *Explosion, a rapid increase in volume and release of energy in an extreme manner *Detonation, an exothermic front accelerating through a medium that eventually drives a shock front Film * ''Blast'' (1997 film), starring Andrew Divoff * ''Blast'' (2000 film), starring Liesel Matthews * ''Blast'' (2004 film), an action comedy film * ''Blast!'' (1972 film) or ''The Final Comedown'', an American drama * ''BLAST!'' (2008 film), a documentary about the BLAST telescope * ''A Blast'', a 2014 film directed by Syllas Tzoumerkas Magazines * ''Blast'' (magazine), a 1914–15 literary magazine of the Vorticist movement * ''Blast'' (U.S. magazine), a 1933–34 American short-story magazine * ''The Blast'' (magazine), a 1916–17 American anarchist periodical Music * Blast (American band), a hardcore punk band * Blast (Russian band), an indie band * ''Blast'' (album), by Holly Johnson, 1989 * ''The Blast'' (album), by Yuvan Shankar Raja, 1999 * "Th ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Sequence Alignment
In bioinformatics, a sequence alignment is a way of arranging the sequences of DNA, RNA, or protein to identify regions of similarity that may be a consequence of functional, structural, or evolutionary relationships between the sequences. Aligned sequences of nucleotide or amino acid residues are typically represented as rows within a matrix. Gaps are inserted between the residues so that identical or similar characters are aligned in successive columns. Sequence alignments are also used for non-biological sequences, such as calculating the distance cost between strings in a natural language or in financial data. Interpretation If two sequences in an alignment share a common ancestor, mismatches can be interpreted as point mutations and gaps as indels (that is, insertion or deletion mutations) introduced in one or both lineages in the time since they diverged from one another. In sequence alignments of proteins, the degree of similarity between amino acids occupying a p ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
POV-Ray
The Persistence of Vision Ray Tracer, most commonly acronymed as POV-Ray, is a cross-platform ray-tracing program that generates images from a text-based scene description. It was originally based on DKBTrace, written by David Kirk Buck and Aaron A. Collins for Amiga computers. There are also influences from the earlier Polyray raytracer because of contributions from its author, Alexander Enzmann. POV-Ray is free and open-source software, with the source code available under the AGPL-3.0-or-later license. History Sometime in the 1980s, David Kirk Buck downloaded the source code for a Unix ray tracer to his Amiga. He experimented with it for a while and eventually decided to write his own ray tracer named DKBTrace after his initials. He posted it to the "You Can Call Me Ray" bulletin board system (BBS) in Chicago, thinking others might be interested in it. In 1987, Aaron A. Collins downloaded DKBTrace and began working on an x86 port of it. He and David Buck collaborated to ad ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Docking (molecular)
In the field of molecular modeling, docking is a method which predicts the preferred orientation of one molecule to a second when a ligand and a target are bound to each other to form a stable complex. Knowledge of the preferred orientation in turn may be used to predict the strength of association or binding affinity between two molecules using, for example, scoring functions. The associations between biologically relevant molecules such as proteins, peptides, nucleic acids, carbohydrates, and lipids play a central role in signal transduction. Furthermore, the relative orientation of the two interacting partners may affect the type of signal produced (e.g., agonism vs antagonism). Therefore, docking is useful for predicting both the strength and type of signal produced. Molecular docking is one of the most frequently used methods in structure-based drug design, due to its ability to predict the binding-conformation of small molecule ligands to the appropriate target b ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |