|
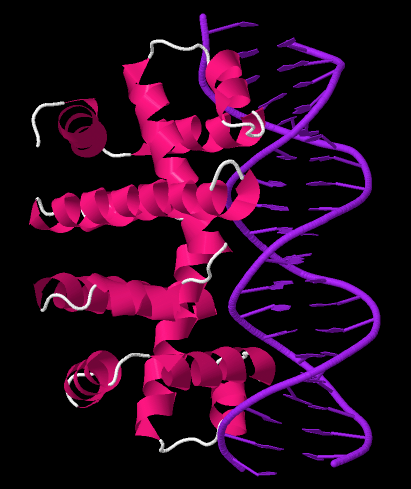
Trp RNA-binding Attenuation Protein
The Tryptophan operon leader is an RNA element found at the 5′ of some bacterial tryptophan operons. The leader sequence can form two different structures known as the terminator and the anti-terminator, based on the Tryptophan amounts in the cell. The leader also codes for very short peptide sequence that is rich in tryptophan. The terminator structure is recognised as a termination signal for RNA polymerase and the operon is not transcribed. This structure forms when the cell has an excess of tryptophan and ribosome movement over the leader transcript is not impeded. When there is a deficiency of the charged tryptophanyl tRNA the ribosome translating the leader peptide stalls and the antiterminator structure can form. This allows RNA polymerase to transcribe the operon. At least 6 different amino acid operons are known to be regulated by this attenuation. Trp RNA-binding attenuation protein The formation of the terminator requires the trp RNA-binding attenuation protein (TR ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Secondary Structure
Protein secondary structure is the three dimensional conformational isomerism, form of ''local segments'' of proteins. The two most common Protein structure#Secondary structure, secondary structural elements are alpha helix, alpha helices and beta sheets, though beta turns and omega loops occur as well. Secondary structure elements typically spontaneously form as an intermediate before the protein protein folding, folds into its three dimensional protein tertiary structure, tertiary structure. Secondary structure is formally defined by the pattern of hydrogen bonds between the Amine, amino hydrogen and carboxyl oxygen atoms in the peptide backbone chain, backbone. Secondary structure may alternatively be defined based on the regular pattern of backbone Dihedral angle#Dihedral angles of proteins, dihedral angles in a particular region of the Ramachandran plot regardless of whether it has the correct hydrogen bonds. The concept of secondary structure was first introduced by Kaj Ulrik ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Sequence Conservation
In evolutionary biology, conserved sequences are identical or similar sequences in nucleic acids ( DNA and RNA) or proteins across species ( orthologous sequences), or within a genome ( paralogous sequences), or between donor and receptor taxa ( xenologous sequences). Conservation indicates that a sequence has been maintained by natural selection. A highly conserved sequence is one that has remained relatively unchanged far back up the phylogenetic tree, and hence far back in geological time. Examples of highly conserved sequences include the RNA components of ribosomes present in all domains of life, the homeobox sequences widespread amongst Eukaryotes, and the tmRNA in Bacteria. The study of sequence conservation overlaps with the fields of genomics, proteomics, evolutionary biology, phylogenetics, bioinformatics and mathematics. History The discovery of the role of DNA in heredity, and observations by Frederick Sanger of variation between animal insulins in 1949, promp ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cis-regulatory Element
''Cis''-regulatory elements (CREs) or ''Cis''-regulatory modules (CRMs) are regions of non-coding DNA which regulate the transcription of neighboring genes. CREs are vital components of genetic regulatory networks, which in turn control morphogenesis, the development of anatomy, and other aspects of embryonic development, studied in evolutionary developmental biology. CREs are found in the vicinity of the genes that they regulate. CREs typically regulate gene transcription by binding to transcription factors. A single transcription factor may bind to many CREs, and hence control the expression of many genes ( pleiotropy). The Latin prefix ''cis'' means "on this side", i.e. on the same molecule of DNA as the gene(s) to be transcribed. CRMs are stretches of DNA, usually 100–1000 DNA base pairs in length, where a number of transcription factors can bind and regulate expression of nearby genes and regulate their transcription rates. They are labeled as ''cis'' because they are ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Bacteria
Bacteria (; singular: bacterium) are ubiquitous, mostly free-living organisms often consisting of one biological cell. They constitute a large domain of prokaryotic microorganisms. Typically a few micrometres in length, bacteria were among the first life forms to appear on Earth, and are present in most of its habitats. Bacteria inhabit soil, water, acidic hot springs, radioactive waste, and the deep biosphere of Earth's crust. Bacteria are vital in many stages of the nutrient cycle by recycling nutrients such as the fixation of nitrogen from the atmosphere. The nutrient cycle includes the decomposition of dead bodies; bacteria are responsible for the putrefaction stage in this process. In the biological communities surrounding hydrothermal vents and cold seeps, extremophile bacteria provide the nutrients needed to sustain life by converting dissolved compounds, such as hydrogen sulphide and methane, to energy. Bacteria also live in symbiotic and parasitic relationsh ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cis-regulatory Element
''Cis''-regulatory elements (CREs) or ''Cis''-regulatory modules (CRMs) are regions of non-coding DNA which regulate the transcription of neighboring genes. CREs are vital components of genetic regulatory networks, which in turn control morphogenesis, the development of anatomy, and other aspects of embryonic development, studied in evolutionary developmental biology. CREs are found in the vicinity of the genes that they regulate. CREs typically regulate gene transcription by binding to transcription factors. A single transcription factor may bind to many CREs, and hence control the expression of many genes ( pleiotropy). The Latin prefix ''cis'' means "on this side", i.e. on the same molecule of DNA as the gene(s) to be transcribed. CRMs are stretches of DNA, usually 100–1000 DNA base pairs in length, where a number of transcription factors can bind and regulate expression of nearby genes and regulate their transcription rates. They are labeled as ''cis'' because they are ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Trp Operon
The ''trp'' operon is a group of genes that are transcribed together, encoding the enzymes that produce the amino acid tryptophan in bacteria. The ''trp'' operon was first characterized in ''Escherichia coli,'' and it has since been discovered in many other bacteria. The operon is regulated so that, when tryptophan is present in the environment, the genes for tryptophan synthesis are repressed. The ''trp'' operon contains five structural genes: ''trpE'', ''trpD'', ''trpC'', ''trpB'', and ''trpA'', which encode the enzymes needed to synthesize tryptophan. It also contains a repressive regulator gene called trpR. When tryptophan is present, the trpR protein binds to the operator, blocking transcription of the ''trp'' operon by RNA polymerase. This operon is an example of repressible negative regulation of gene expression. The repressor protein binds to the operator in the presence of tryptophan (repressing transcription) and is released from the operon when tryptophan is absent ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Tryptophan
Tryptophan (symbol Trp or W) is an α-amino acid that is used in the biosynthesis of proteins. Tryptophan contains an α-amino group, an α- carboxylic acid group, and a side chain indole, making it a polar molecule with a non-polar aromatic beta carbon substituent. It is essential in humans, meaning that the body cannot synthesize it and it must be obtained from the diet. Tryptophan is also a precursor to the neurotransmitter serotonin, the hormone melatonin, and vitamin B3. It is encoded by the codon UGG. Like other amino acids, tryptophan is a zwitterion at physiological pH where the amino group is protonated (–; pKa = 9.39) and the carboxylic acid is deprotonated ( –COO−; pKa = 2.38). Humans and many animals cannot synthesize tryptophan: they need to obtain it through their diet, making it an essential amino acid. Function Amino acids, including tryptophan, are used as building blocks in protein biosynthesis, and proteins are required to sustain life. Man ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
RNA Polymerase
In molecular biology, RNA polymerase (abbreviated RNAP or RNApol), or more specifically DNA-directed/dependent RNA polymerase (DdRP), is an enzyme that synthesizes RNA from a DNA template. Using the enzyme helicase, RNAP locally opens the double-stranded DNA so that one strand of the exposed nucleotides can be used as a template for the synthesis of RNA, a process called transcription. A transcription factor and its associated transcription mediator complex must be attached to a DNA binding site called a promoter region before RNAP can initiate the DNA unwinding at that position. RNAP not only initiates RNA transcription, it also guides the nucleotides into position, facilitates attachment and elongation, has intrinsic proofreading and replacement capabilities, and termination recognition capability. In eukaryotes, RNAP can build chains as long as 2.4 million nucleotides. RNAP produces RNA that, functionally, is either for protein coding, i.e. messenger RNA (mRNA); or n ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ribosome
Ribosomes ( ) are macromolecular machines, found within all cells, that perform biological protein synthesis (mRNA translation). Ribosomes link amino acids together in the order specified by the codons of messenger RNA (mRNA) molecules to form polypeptide chains. Ribosomes consist of two major components: the small and large ribosomal subunits. Each subunit consists of one or more ribosomal RNA (rRNA) molecules and many ribosomal proteins (RPs or r-proteins). The ribosomes and associated molecules are also known as the ''translational apparatus''. Overview The sequence of DNA that encodes the sequence of the amino acids in a protein is transcribed into a messenger RNA chain. Ribosomes bind to messenger RNAs and use their sequences for determining the correct sequence of amino acids to generate a given protein. Amino acids are selected and carried to the ribosome by transfer RNA (tRNA) molecules, which enter the ribosome and bind to the messenger RNA chain via an anti-c ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
TRNA
Transfer RNA (abbreviated tRNA and formerly referred to as sRNA, for soluble RNA) is an adaptor molecule composed of RNA, typically 76 to 90 nucleotides in length (in eukaryotes), that serves as the physical link between the mRNA and the amino acid sequence of proteins. tRNAs genes from Bacteria are typically shorter (mean = 77.6 bp) than tRNAs from Archaea (mean = 83.1 bp) and eukaryotes (mean = 84.7 bp). The mature tRNA follows an opposite pattern with tRNAs from Bacteria being usually longer (median = 77.6 nt) than tRNAs from Archaea (median = 76.8 nt), with eukaryotes exhibiting the shortest mature tRNAs (median = 74.5 nt). Transfer RNA (tRNA) does this by carrying an amino acid to the protein synthesizing machinery of a cell called the ribosome. Complementation of a 3-nucleotide codon in a messenger RNA (mRNA) by a 3-nucleotide anticodon of the tRNA results in protein synthesis based on the mRNA code. As such, tRNAs are a necessary component of translation, the biological ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
2zd0
Zd or ZD may refer to: * Nissan ZD engine, an automobile engine * Zero Defects, a management program to eliminate defects in industrial production, popular in American industry in the late 1960s * Zero-defects mentality, in which a command-and-control structure does not tolerate mistakes * Zertifikat Deutsch, a certificate of German language ability * Ziff Davis Ziff Davis, Inc. is an American digital media and internet company. First founded in 1927 by William Bernard Ziff Sr. and Bernard George Davis, the company primarily owns technology-oriented media websites, online shopping-related services, an ..., a magazine publisher and Internet information provider * Zooey Deschanel, an American actress and musician {{disambiguation ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein
Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residues. Proteins perform a vast array of functions within organisms, including catalysing metabolic reactions, DNA replication, responding to stimuli, providing structure to cells and organisms, and transporting molecules from one location to another. Proteins differ from one another primarily in their sequence of amino acids, which is dictated by the nucleotide sequence of their genes, and which usually results in protein folding into a specific 3D structure that determines its activity. A linear chain of amino acid residues is called a polypeptide. A protein contains at least one long polypeptide. Short polypeptides, containing less than 20–30 residues, are rarely considered to be proteins and are commonly called peptides. The individual amino acid residues are bonded together by peptide bonds and adjacent amino acid residues. The sequence of amino acid residue ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |