|
Tandem Affinity Purification
Tandem affinity purification (TAP) is an immunoprecipitation-based purification technique for studying protein–protein interactions. The goal is to extract from a cell only the protein of interest, in complex with any other proteins it interacted with. TAP uses two types of agarose beads that bind to the protein of interest and that can be separated from the cell lysate by centrifugation, without disturbing, denaturing or contaminating the involved complexes. To enable the protein of interest to bind to the beads, it is tagged with a designed piece, the TAP tag. The original TAP method involves the fusion of the TAP tag to the C-terminus of the protein under study. The TAP tag consists of three components: a calmodulin binding peptide (CBP), TEV protease cleavage site, and two Protein A domains, which bind tightly to IgG (making a TAP tag a type of epitope tag). Many other tag/bead/eluent combinations have been proposed since the TAP principle was first published. Variant ta ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Plasmids
A plasmid is a small, extrachromosomal DNA molecule within a cell that is physically separated from chromosomal DNA and can replicate independently. They are most commonly found as small circular, double-stranded DNA molecules in bacteria; however, plasmids are sometimes present in archaea and eukaryotic organisms. In nature, plasmids often carry genes that benefit the survival of the organism and confer selective advantage such as antibiotic resistance. While chromosomes are large and contain all the essential genetic information for living under normal conditions, plasmids are usually very small and contain only additional genes that may be useful in certain situations or conditions. Artificial plasmids are widely used as vectors in molecular cloning, serving to drive the replication of recombinant DNA sequences within host organisms. In the laboratory, plasmids may be introduced into a cell via transformation. Synthetic plasmids are available for procurement over the intern ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Proteomics
Proteomics is the large-scale study of proteins. Proteins are vital parts of living organisms, with many functions such as the formation of structural fibers of muscle tissue, enzymatic digestion of food, or synthesis and replication of DNA. In addition, other kinds of proteins include antibodies that protect an organism from infection, and hormones that send important signals throughout the body. The proteome is the entire set of proteins produced or modified by an organism or system. Proteomics enables the identification of ever-increasing numbers of proteins. This varies with time and distinct requirements, or stresses, that a cell or organism undergoes. Proteomics is an interdisciplinary domain that has benefited greatly from the genetic information of various genome projects, including the Human Genome Project. It covers the exploration of proteomes from the overall level of protein composition, structure, and activity, and is an important component of functional genomics. ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Photo-reactive Amino Acid Analog
Photo-reactive amino acid analogs are artificial analogs of natural amino acids that can be used for crosslinking of protein complexes. Photo-reactive amino acid analogs may be incorporated into proteins and peptides ''in vivo'' or in ''vitro''. Photo-reactive amino acid analogs in common use are photoreactive diazirine analogs to leucine and methionine, and ''para''-benzoylphenylalanine. Upon exposure to ultraviolet light, they are activated and covalently bind to interacting proteins that are within a few angstroms of the photo-reactive amino acid analog. L-Photo-leucine and L-photo-methionine are analogs of the naturally occurring L-leucine and L-methionine amino acids that are endogenously incorporated into the primary sequence of proteins during synthesis using the normal translation machinery. They are then ultraviolet light (UV)-activated to covalently crosslink proteins within protein–protein interaction domains in their native ''in-vivo'' environment. The method enables ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Yeast Two-hybrid
Two-hybrid screening (originally known as yeast two-hybrid system or Y2H) is a molecular biology technique used to discover protein–protein interactions (PPIs) and protein–DNA interactions by testing for physical interactions (such as binding) between two proteins or a single protein and a DNA molecule, respectively. The premise behind the test is the activation of downstream reporter gene(s) by the binding of a transcription factor onto an upstream activating sequence (UAS). For two-hybrid screening, the transcription factor is split into two separate fragments, called the DNA-binding domain (DBD or often also abbreviated as BD) and activating domain (AD). The BD is the domain responsible for binding to the UAS and the AD is the domain responsible for the activation of transcription. The Y2H is thus a protein-fragment complementation assay. History Pioneered by Stanley Fields and Ok-Kyu Song in 1989, the technique was originally designed to detect protein–protein ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Transient Protein Interactions
ECHELON, originally a secret government code name, is a surveillance program (signals intelligence/SIGINT collection and analysis network) operated by the five signatory states to the UKUSA Security Agreement:Given the 5 dialects that use the terms, UKUSA can be pronounced from "You-Q-SA" to "Oo-Coo-SA", AUSCANNZUKUS can be pronounced from "Oz-Can-Zuke-Us" to "Orse-Can-Zoo-Cuss". :From Talk:UKUSA Agreement: "Per documents officially released by both the Government Communications Headquarters and the National Security Agency, this agreement is referred to as the UKUSA Agreement. This name is subsequently used by media sources reporting on the story, as written in new references used for the article. The NSA press release provides a pronunciation guide, indicating that "UKUSA" should not be read as two separate entities.(National Security Agency)" Australia, Canada, New Zealand, the United Kingdom and the United States, also known as the Five Eyes. Created in the late 1960s ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
In Vivo
Studies that are ''in vivo'' (Latin for "within the living"; often not italicized in English) are those in which the effects of various biological entities are tested on whole, living organisms or cells, usually animals, including humans, and plants, as opposed to a tissue extract or dead organism. This is not to be confused with experiments done ''in vitro'' ("within the glass"), i.e., in a laboratory environment using test tubes, Petri dishes, etc. Examples of investigations ''in vivo'' include: the pathogenesis of disease by comparing the effects of bacterial infection with the effects of purified bacterial toxins; the development of non-antibiotics, antiviral drugs, and new drugs generally; and new surgical procedures. Consequently, animal testing and clinical trials are major elements of ''in vivo'' research. ''In vivo'' testing is often employed over ''in vitro'' because it is better suited for observing the overall effects of an experiment on a living subject. In dr ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Mass Spectrometry
Mass spectrometry (MS) is an analytical technique that is used to measure the mass-to-charge ratio of ions. The results are presented as a ''mass spectrum'', a plot of intensity as a function of the mass-to-charge ratio. Mass spectrometry is used in many different fields and is applied to pure samples as well as complex mixtures. A mass spectrum is a type of plot of the ion signal as a function of the mass-to-charge ratio. These spectra are used to determine the elemental or isotopic signature of a sample, the masses of particles and of molecules, and to elucidate the chemical identity or structure of molecules and other chemical compounds. In a typical MS procedure, a sample, which may be solid, liquid, or gaseous, is ionized, for example by bombarding it with a beam of electrons. This may cause some of the sample's molecules to break up into positively charged fragments or simply become positively charged without fragmenting. These ions (fragments) are then separated accordin ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
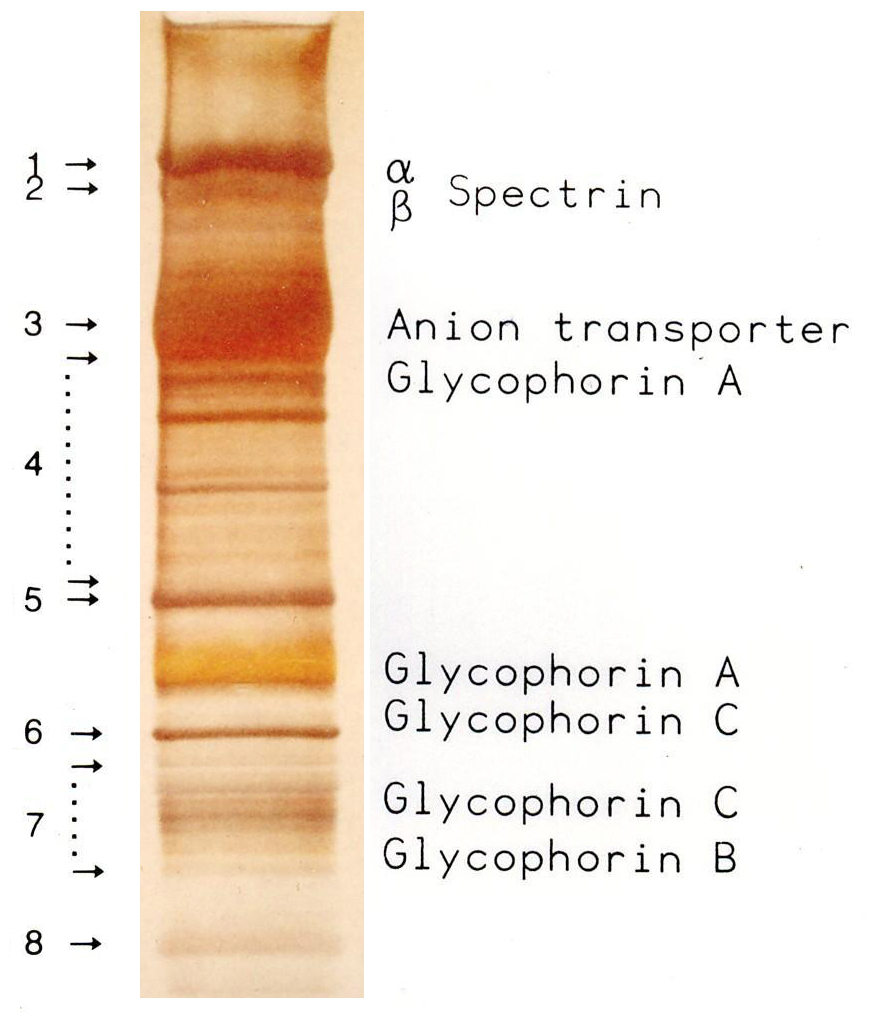
SDS-PAGE
SDS-PAGE (sodium dodecyl sulfate–polyacrylamide gel electrophoresis) is a discontinuous electrophoretic system developed by Ulrich K. Laemmli which is commonly used as a method to separate proteins with molecular masses between 5 and 250 kDa. The combined use of sodium dodecyl sulfate (SDS, also known as sodium lauryl sulfate) and polyacrylamide gel allows to eliminate the influence of structure and charge, and proteins are separated solely on the basis of differences in their molecular weight. Properties SDS-PAGE is an electrophoresis method that allows protein separation by mass. The medium (also referred to as ′matrix′) is a polyacrylamide-based discontinuous gel. The polyacrylamide-gel is typically sandwiched between two glass plates in a slab gel. Although tube gels (in glass cylinders) were used historically, they were rapidly made obsolete with the invention of the more convenient slab gels. In addition, SDS (sodium dodecyl sulfate) is used. About 1.4 grams of S ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
EGTA (chemical)
EGTA (ethylene glycol-bis(β-aminoethyl ether)-''N'',''N'',''N''′,''N''′-tetraacetic acid), also known as egtazic acid (INN, USAN), is an aminopolycarboxylic acid, a chelating agent. It is a white solid that is related to the better known EDTA. Compared to EDTA, it has a lower affinity for magnesium, making it more selective for calcium ions. It is useful in buffer solutions that resemble the environment in living cells where calcium ions are usually at least a thousandfold less concentrated than magnesium. The p''K''a for binding of calcium ions by tetrabasic EGTA is 11.00, but the protonated forms do not significantly contribute to binding, so at pH 7, the apparent p''K''a becomes 6.91. See Qin ''et al.'' for an example of a p''K''a calculation. EGTA has also been used experimentally for the treatment of animals with cerium poisoning and for the separation of thorium from the mineral monazite. EGTA is used as a compound in elution buffer in the protein purification te ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |

.jpg)

.jpg)