|
Trithorax-group Proteins
Trithorax-group proteins (TrxG) are a heterogeneous collection of proteins whose main action is to maintain gene expression. They can be categorized into three general classes based on molecular function: # histone-modifying TrxG proteins # chromatin-remodeling TrxG proteins # DNA-binding TrxG proteins, plus other TrxG proteins not categorized in the first three classes. Discovery The founding member of TrxG proteins, trithorax (trx), was discovered ~1978 by Philip Ingham as part of his doctoral thesis while a graduate student in the laboratory of J.R.S. Whittle at the University of Sussex. Histone-lysine ''N''-methyltransferase 2A is the human homolog of trx. The table contains names of Drosophila TrxG members. Homologs in other species may have different names. Function Trithorax-group proteins typically function in large complexes formed with other proteins. The complexes formed by TrxG proteins are divided into two groups: histone-modifying complexes and ATP-depende ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Gene Expression
Gene expression is the process by which information from a gene is used in the synthesis of a functional gene product that enables it to produce end products, protein or non-coding RNA, and ultimately affect a phenotype, as the final effect. These products are often proteins, but in non-protein-coding genes such as transfer RNA (tRNA) and small nuclear RNA (snRNA), the product is a functional non-coding RNA. Gene expression is summarized in the central dogma of molecular biology first formulated by Francis Crick in 1958, further developed in his 1970 article, and expanded by the subsequent discoveries of reverse transcription and RNA replication. The process of gene expression is used by all known life—eukaryotes (including multicellular organisms), prokaryotes (bacteria and archaea), and utilized by viruses—to generate the macromolecular machinery for life. In genetics, gene expression is the most fundamental level at which the genotype gives rise to the phenotype, '' ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Histone H4
Histone H4 is one of the five main histone proteins involved in the structure of chromatin in eukaryote, eukaryotic cells. Featuring a main globular domain and a long N-terminus, N-terminal tail, H4 is involved with the structure of the nucleosome of the 'beads on a string' organization. Histone proteins are highly post-translationally modified. Covalently bonded modifications include acetylation and methylation of the N-terminal tails. These modifications may alter Gene expression, expression of genes located on DNA associated with its parent histone octamer. Histone H4 is an important protein in the structure and function of chromatin, where its sequence variants and variable modification states are thought to play a role in the dynamic and long term regulation of genes. Genetics Histone H4 is encoded in multiple genes at different loci including: HIST1H4A, HIST1H4B, HIST1H4C, HIST1H4D, HIST1H4E, HIST1H4F, HIST1H4G, HIST1H4H, HIST1H4I, HIST1H4J, HIST1H4K, HIST1H4L, HIST2H4A, H ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DNA-binding Proteins
DNA-binding proteins are proteins that have DNA-binding domains and thus have a specific or general affinity for single- or double-stranded DNA. Sequence-specific DNA-binding proteins generally interact with the major groove of B-DNA, because it exposes more functional groups that identify a base pair. However, there are some known minor groove DNA-binding ligands such as netropsin, distamycin, Hoechst 33258, pentamidine, DAPI and others. Examples DNA-binding proteins include transcription factors which modulate the process of transcription, various polymerases, nucleases which cleave DNA molecules, and histones which are involved in chromosome packaging and transcription in the cell nucleus. DNA-binding proteins can incorporate such domains as the zinc finger, the helix-turn-helix, and the leucine zipper (among many others) that facilitate binding to nucleic acid. There are also more unusual examples such as transcription activator like effectors. Non-specific DNA-protein ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
PRMT4 Pathway
Protein arginine N-methyltransferase-4 (PRMT4/CARM1) methylation of arginine residues within proteins plays a critical key role in transcriptional regulation (see the PRMT4 pathway on the left). PRMT4 binds to the classes of transcriptional activators known as p160 and CBP/p300.Zika E, et al., Interplay among coactivator-associated arginine methyltransferase 1, CBP, and CIITA in IFN-gamma-inducible MHC-II gene expression. Proc Natl Acad Sci U S A. 2005 Nov 8;102(45):16321-6 The modified forms of these proteins are involved in stimulation of gene expression via steroid hormone receptors. Significantly, PRMT4 methylates core histones H3 and H4, which are also targets of the histone acetylase activity of CBP/p300 coactivators. PRMT4 recruitment of chromatin by binding to coactivators increases histone methylation and enhances the accessibility of promoter regions for transcription. Methylation of the transcriptional coactivator CBP by PRMT4 inhibits binding to CREB and thereby partiti ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
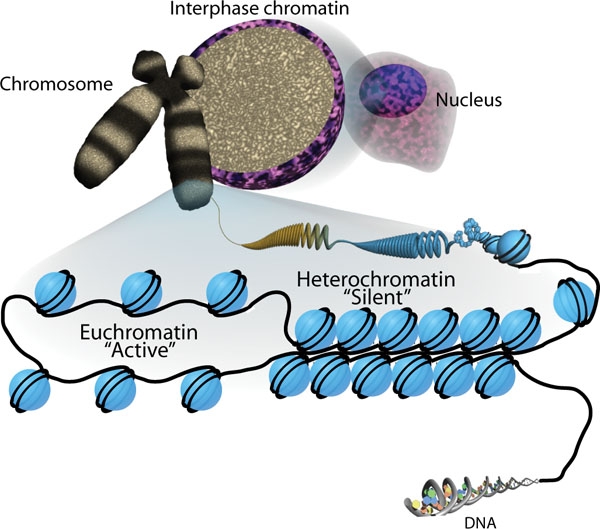
Nucleosome
A nucleosome is the basic structural unit of DNA packaging in eukaryotes. The structure of a nucleosome consists of a segment of DNA wound around eight histone proteins and resembles thread wrapped around a spool. The nucleosome is the fundamental subunit of chromatin. Each nucleosome is composed of a little less than two turns of DNA wrapped around a set of eight proteins called histones, which are known as a histone octamer. Each histone octamer is composed of two copies each of the histone proteins H2A, H2B, H3, and H4. DNA must be compacted into nucleosomes to fit within the cell nucleus. In addition to nucleosome wrapping, eukaryotic chromatin is further compacted by being folded into a series of more complex structures, eventually forming a chromosome. Each human cell contains about 30 million nucleosomes. Nucleosomes are thought to carry epigenetically inherited information in the form of covalent modifications of their core histones. Nucleosome positions in the gen ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Histone-Modifying Enzymes
Histone-modifying enzymes are enzymes involved in the modification of histone substrates after protein translation and affect cellular processes including gene expression. To safely store the eukaryotic genome, DNA is wrapped around four core histone proteins (H3, H4, H2A, H2B), which then join to form nucleosomes. These nucleosomes further fold together into highly condensed chromatin, which renders the organism's genetic material far less accessible to the factors required for gene transcription, DNA replication, recombination and repair. Subsequently, eukaryotic organisms have developed intricate mechanisms to overcome this repressive barrier imposed by the chromatin through histone modification, a type of post-translational modification which typically involves covalently attaching certain groups to histone residues. Once added to the histone, these groups (directly or indirectly) elicit either a loose and open histone conformation, euchromatin, or a tight and closed histone ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Histone Methyltransferase
Histone methyltransferases (HMT) are histone-modifying enzymes (e.g., histone-lysine N-methyltransferases and histone-arginine N-methyltransferases), that catalyze the transfer of one, two, or three methyl groups to lysine and arginine residues of histone proteins. The attachment of methyl groups occurs predominantly at specific lysine or arginine residues on histones H3 and H4. Two major types of histone methyltranferases exist, lysine-specific (which can be SET (Su(var)3-9, Enhancer of Zeste, Trithorax) domain containing or non-SET domain containing) and arginine-specific. In both types of histone methyltransferases, S-Adenosyl methionine (SAM) serves as a cofactor and methyl donor group. The genomic DNA of eukaryotes associates with histones to form chromatin. The level of chromatin compaction depends heavily on histone methylation and other post-translational modifications of histones. Histone methylation is a principal epigenetic modification of chromatin that determines ge ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Histone Deacetylases
Histone deacetylases (, HDAC) are a class of enzymes that remove acetyl groups (O=C-CH3) from an ε-N-acetyl lysine amino acid on a histone, allowing the histones to wrap the DNA more tightly. This is important because DNA is wrapped around histones, and DNA expression is regulated by acetylation and de-acetylation. Its action is opposite to that of histone acetyltransferase. HDAC proteins are now also called lysine deacetylases (KDAC), to describe their function rather than their target, which also includes non-histone proteins. HDAC super family Together with the acetylpolyamine amidohydrolases and the acetoin utilization proteins, the histone deacetylases form an ancient protein superfamily known as the histone deacetylase superfamily. Classes of HDACs in higher eukaryotes HDACs, are classified in four classes depending on sequence homology to the yeast original enzymes and domain organization: HDAC (except class III) contain zinc and are known as Zn2+-dependent hi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Histone Acetyltransferase
Histone acetyltransferases (HATs) are enzymes that acetylate conserved lysine amino acids on histone proteins by transferring an acetyl group from acetyl-CoA to form ε-''N''-acetyllysine. DNA is wrapped around histones, and, by transferring an acetyl group to the histones, genes can be turned on and off. In general, histone acetylation increases gene expression. In general, histone acetylation is linked to transcriptional activation and associated with euchromatin. Euchromatin, which is less densely compact, allows transcription factors to bind more easily to regulatory sites on DNA, causing transcriptional activation. When it was first discovered, it was thought that acetylation of lysine neutralizes the positive charge normally present, thus reducing affinity between histone and (negatively charged) DNA, which renders DNA more accessible to transcription factors. Research has emerged, since, to show that lysine acetylation and other posttranslational modifications of hist ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
HIstome
HIstome is a database that provides information about human histone proteins, their sites of modifications, variants and modifying enzymes, and diseases linked to histone modifications. Update HISTome2 is a updated version of the HIstome database released in 2020. See also * Histone In biology, histones are highly basic proteins abundant in lysine and arginine residues that are found in eukaryotic cell nuclei. They act as spools around which DNA winds to create structural units called nucleosomes. Nucleosomes in turn a ... References External links * http://www.iiserpune.ac.in/~coee/histome/ Biological databases Epigenetics {{Biodatabase-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Embryogenesis
An embryo is an initial stage of development of a multicellular organism. In organisms that reproduce sexually, embryonic development is the part of the life cycle that begins just after fertilization of the female egg cell by the male sperm cell. The resulting fusion of these two cells produces a single-celled zygote that undergoes many cell divisions that produce cells known as blastomeres. The blastomeres are arranged as a solid ball that when reaching a certain size, called a morula, takes in fluid to create a cavity called a blastocoel. The structure is then termed a blastula, or a blastocyst in mammals. The mammalian blastocyst hatches before implantating into the endometrial lining of the womb. Once implanted the embryo will continue its development through the next stages of gastrulation, neurulation, and organogenesis. Gastrulation is the formation of the three germ layers that will form all of the different parts of the body. Neurulation forms the nervous sys ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
X-chromosome Inactivation
X-inactivation (also called Lyonization, after English geneticist Mary Lyon) is a process by which one of the copies of the X chromosome is inactivated in therian female mammals. The inactive X chromosome is silenced by being packaged into a transcriptionally inactive structure called heterochromatin. As nearly all female mammals have two X chromosomes, X-inactivation prevents them from having twice as many X chromosome gene products as males, who only possess a single copy of the X chromosome (see dosage compensation). The choice of which X chromosome will be inactivated in a particular embryonic cell is random in placental mammals such as humans, but once an X chromosome is inactivated it will remain inactive throughout the lifetime of the cell and its descendants in the organism (its cell line). The result is that the choice of inactivated X chromosome in all the cells of the organism is a random distribution, often with about half the cells having the paternal X chromos ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |