|
T-REX (webserver)
T-REX (Tree and Reticulogram Reconstruction) is a freely available web server, developed at the department of Computer Science of the Université du Québec à Montréal, dedicated to the inference, validation and visualization of phylogenetic trees and phylogenetic networks. The T-REX web server allows the users to perform several popular methods of phylogenetic analysis as well as some new phylogenetic applications for inferring, drawing and validating phylogenetic trees and networks. Phylogenetic inference The following methods for inferring and validating phylogenetic trees using distances are available: Neighbor joining (NJ),NINJA large-scale Neighbor Joining''k'') = exp(-''k'')), and the constant a is a tuning factor accounting for the deviation intensity (as described in Guindon and Gascuel (2002), the value of a was set to 0.8). The random trees generated by this procedure have depth of O(log (''n'')). References {{Reflist External links Official T-REX Web server page ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Web Server
A web server is computer software and underlying Computer hardware, hardware that accepts requests via Hypertext Transfer Protocol, HTTP (the network protocol created to distribute web content) or its secure variant HTTPS. A user agent, commonly a web browser or web crawler, initiates communication by making a request for a web page or other Web Resource, resource using HTTP, and the server (computing), server responds with the content of that resource or an List of HTTP status codes, error message. A web server can also accept and store resources sent from the user agent if configured to do so. The hardware used to run a web server can vary according to the volume of requests that it needs to handle. At the low end of the range are embedded systems, such as a router (computing), router that runs a small web server as its configuration interface. A high-traffic Internet website might handle requests with hundreds of servers that run on racks of high-speed computers. A reso ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
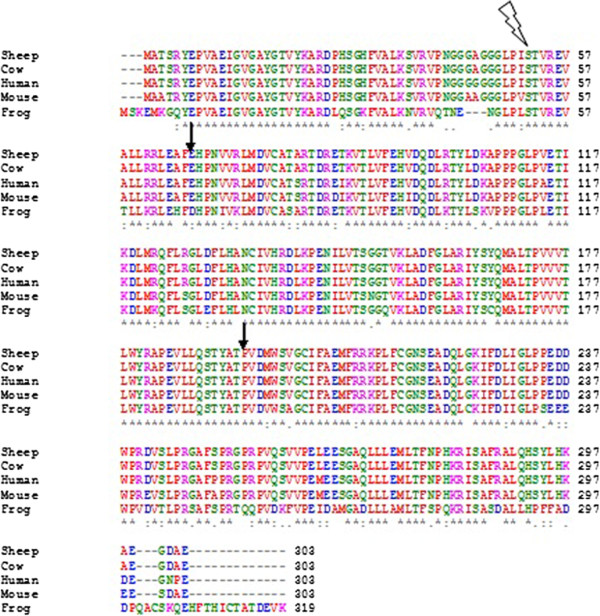
MAFFT
In bioinformatics, MAFFT (multiple alignment using fast Fourier transform) is a program used to create multiple sequence alignments of amino acid or nucleotide sequences. Published in 2002, the first version used an algorithm based on multiple sequence alignment#Progressive alignment construction, progressive alignment, in which the sequences were clustered with the help of the fast Fourier transform. Subsequent versions of MAFFT have added other algorithms and modes of operation, including options for faster alignment of large numbers of sequences, higher accuracy alignments, alignment of non-coding RNA sequences, and the addition of new sequences to existing alignments. History There have been many variations of the MAFFT software, some of which are listed below: * ''MAFFT'' – The first version, created by Kazutaka Katoh in 2002, used an algorithm based on multiple sequence alignment#Progressive alignment construction, progressive alignment, in which the sequences were cluste ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Distance Matrix
In mathematics, computer science and especially graph theory, a distance matrix is a square matrix (two-dimensional array) containing the distances, taken pairwise, between the elements of a set. Depending upon the application involved, the ''distance'' being used to define this matrix may or may not be a metric (mathematics), metric. If there are elements, this matrix will have size . In graph-theoretic applications, the elements are more often referred to as points, nodes or vertices. Non-metric distance matrix In general, a distance matrix is a weighted adjacency matrix of some graph. In a Network (mathematics), network, a directed graph with weights assigned to the arcs, the distance between two nodes of the network can be defined as the minimum of the sums of the weights on the shortest paths joining the two nodes (where the number of steps in the path is bounded). This distance function, while well defined, is not a metric. There need be no restrictions on the weights oth ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Newick
Newick is a village, civil parish and electoral ward in the Lewes District of East Sussex, England. It is located on the A272 road east of Haywards Heath. The parish church, St. Mary's, dates mainly from the Victorian era, but still has a Norman window. Zion Chapel, a Strict Baptist chapel, was built in 1834 and converted to flats in 2001. Newick Evangelical Free Church, originally a mission hall, opened in 1892. The village is home to three pubs (''The Crown Inn'', ''The Royal Oak'', and ''The Bull Inn''), one restaurant (Newick Tandoori), a butcher, a baker, a pharmacy, as well as a number of other businesses. There is also a primary school, a health centre, a village hall known originally as the 'Derek Hall', and a post office. Like many other places in Sussex, Newick holds an annual Bonfire Night celebration on the Saturday before Lewes Bonfire Night. Many of the local bonfire societies join the procession. History Newick was a dispersed settlement until the Second W ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Robinson–Foulds Metric
The Robinson–Foulds or symmetric difference metric, often abbreviated as the RF distance, is a simple way to calculate the distance between phylogenetic trees. It is defined as ( + ) where is the number of partitions of data implied by the first tree but not the second tree and is the number of partitions of data implied by the second tree but not the first tree (although some software implementations divide the RF metric by 2 and others scale the RF distance to have a maximum value of 1). The partitions are calculated for each tree by removing each branch. Thus, the number of eligible partitions for each tree is equal to the number of branches in that tree. RF distances have been criticized as biased, but they represent a relatively intuitive measure of the distances between phylogenetic trees and therefore remain widely used (the original 1981 paper describing Robinson-Foulds distances was cited more than 2700 times by 2023 based on Google Scholar). Nevertheless, the biases i ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Multiple Sequence Alignment
Multiple sequence alignment (MSA) is the process or the result of sequence alignment of three or more biological sequences, generally protein, DNA, or RNA. These alignments are used to infer evolutionary relationships via phylogenetic analysis and can highlight homologous features between sequences. Alignments highlight mutation events such as point mutations (single amino acid or nucleotide changes), insertion mutations and deletion mutations, and alignments are used to assess sequence conservation and infer the presence and activity of protein domains, tertiary structures, secondary structures, and individual amino acids or nucleotides. Multiple sequence alignments require more sophisticated methodologies than pairwise alignments, as they are more computationally complex. Most multiple sequence alignment programs use heuristic methods rather than global optimization because identifying the optimal alignment between more than a few sequences of moderate length is prohibiti ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
ClustalW
Clustal is a computer program used for multiple sequence alignment in bioinformatics. The software and its algorithms have gone through several iterations, with ClustalΩ (Omega) being the latest version . It is available as standalone software, via a web interface, and through a server hosted by the European Bioinformatics Institute. Clustal has been an important bioinformatic software, with two of its academic publications amongst the top 100 papers cited of all time, according to Nature in 2014. History Version history * Clustal: The original software for multiple sequence alignments, created by Des Higgins in 1988, was based on deriving a guide tree from pairwise sequences of amino acids or nucleotides. * ClustalV: The second generation of Clustal, released in 1992. It introduced the ability to create new alignments from existing alignments in a process known as phylogenetic tree reconstruction. ClustalV also added the option to create trees using the neighbor joini ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
MUSCLE (alignment Software)
MUltiple Sequence Comparison by Log-Expectation (MUSCLE) is a computer software for multiple sequence alignment of protein and nucleotide sequences. It is Software license, licensed as public domain. The method was published by Robert C. Edgar in two papers in 2004. The first paper, published in ''Nucleic Acids Research'', introduced the sequence alignment algorithm. The second paper, published in ''BMC Bioinformatics'', presented more technical details. MUSCLE up to version 3 uses a progressive-refinement method. Since version 5 it uses a hidden Markov model similar to ProbCons. History Robert C. Edgar Edgar graduated in 1982 from University College London, BSc in Physics, PhD in Particle physics. He pursued software development post-graduation and founded his own company, Parity Software, in 1988. In 2001, he began working with coding algorithms after attending a seminar at the University of California Berkley. From 2001-present day Edgar has contributed to or been the sole ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Horizontal Gene Transfer
Horizontal gene transfer (HGT) or lateral gene transfer (LGT) is the movement of genetic material between organisms other than by the ("vertical") transmission of DNA from parent to offspring (reproduction). HGT is an important factor in the evolution of many organisms. HGT is influencing scientific understanding of higher-order evolution while more significantly shifting perspectives on bacterial evolution. Horizontal gene transfer is the primary mechanism for the spread of antibiotic resistance in bacteria, and plays an important role in the evolution of bacteria that can degrade novel compounds such as human-created Bactericide, pesticides and in the evolution, maintenance, and transmission of virulence. It often involves Temperateness (virology), temperate bacteriophages and plasmids. Genes responsible for antibiotic resistance in one species of bacteria can be transferred to another species of bacteria through various mechanisms of HGT such as Transformation (genetics), tr ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Université Du Québec à Montréal
The (UQAM; ), is a French language, French-language public university, public research university based in Montreal, Quebec, Canada. It is the largest constituent element of the system. UQAM was founded on April 9, 1969, by the government of Quebec, through the merger of the , a fine arts school; the , a classical college; and a number of smaller schools. Although part of the UQ network, UQAM possesses a relative independence which allows it to choose its rector. In the fall of 2018, the university welcomed some 40,738 students, including 3,859 international students from 95 countries, in a total of 310 distinct programs of study, managed by six faculties (Arts, Education, Communication, Political Science and Law, Science and Social science) and one school (Management). It offers Bachelor's degree, Bachelors, Master's degree, Masters, and Doctor of Philosophy, Doctoral degrees. It is one of Montreal's two French-language universities, along with the , and only 1% of its stud ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
FASTA Format
In bioinformatics and biochemistry, the FASTA format is a text-based format for representing either nucleotide sequences or amino acid (protein) sequences, in which nucleotides or amino acids are represented using single-letter codes. The format allows for sequence names and comments to precede the sequences. It originated from the FASTA software package and has since become a near-universal standard in bioinformatics. The simplicity of FASTA format makes it easy to manipulate and parse sequences using text-processing tools and scripting languages. Overview A sequence begins with a greater-than character (">") followed by a description of the sequence (all in a single line). The lines immediately following the description line are the sequence representation, with one letter per amino acid or nucleic acid, and are typically no more than 80 characters in length. For example: >MCHU - Calmodulin - Human, rabbit, bovine, rat, and chicken MADQLTEEQIAEFKEAFSLFDKDGDGTITTKELGTV ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Newick Format
In mathematics and phylogenetics, Newick tree format (or Newick notation or New Hampshire tree format) is a way of representing graph-theoretical trees with edge lengths using parentheses and commas. It was adopted by James Archie, William H. E. Day, Joseph Felsenstein, Wayne Maddison, Christopher Meacham, F. James Rohlf, and David Swofford, at two meetings in 1986, the second of which was at Newick's restaurant in Dover, New Hampshire, US. The adopted format is a generalization of the format developed by Meacham in 1984 for the first tree-drawing programs in Felsenstein's PHYLIP package. Examples The following tree: could be represented in Newick format in several ways ((,)); ''no nodes are named'' (A,B,(C,D)); ''leaf nodes are named'' (A,B,(C,D)E)F; ''all nodes are named'' (:0.1,:0.2,(:0.3,:0.4):0.5); ''all but root node have a distance to parent'' (:0.1,:0.2,(:0.3,:0.4):0.5): ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |