|
Synthesis (cell Cycle)
S phase (Synthesis Phase) is the phase of the cell cycle in which DNA is DNA replication, replicated, occurring between G1 phase, G1 phase and G2 phase, G2 phase. Since accurate duplication of the genome is critical to successful cell division, the processes that occur during S-phase are tightly regulated and widely conserved. Regulation Entry into S-phase is controlled by the G1 restriction point (R), which commits cells to the remainder of the cell-cycle if there is adequate nutrients and growth signaling. This transition is essentially irreversible; after passing the restriction point, the cell will progress through S-phase even if environmental conditions become unfavorable. Accordingly, entry into S-phase is controlled by molecular pathways that facilitate a rapid, unidirectional shift in cell state. In yeast, for instance, cell growth induces accumulation of Cln3 cyclin, which complexes with the Cyclin-dependent kinase, cyclin dependent kinase CDK2. The Cln3-CDK2 complex ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Asymmetry In The Synthesis Of Leading And Lagging Strands
Asymmetry is the absence of, or a violation of, symmetry (the property of an object being invariant to a transformation, such as reflection). Symmetry is an important property of both physical and abstract systems and it may be displayed in precise terms or in more aesthetic terms. The absence of or violation of symmetry that are either expected or desired can have important consequences for a system. In organisms Due to how cell (biology), cells divide in organisms, asymmetry in organisms is fairly usual in at least one dimension, with symmetry (biology), biological symmetry also being common in at least one dimension. Louis Pasteur proposed that biological molecules are asymmetric because the cosmic [i.e. physical] forces that preside over their formation are themselves asymmetric. While at his time, and even now, the symmetry of physical processes are highlighted, it is known that there are fundamental physical asymmetries, starting with time. Asymmetry in biology Asym ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Proliferating Cell Nuclear Antigen
Proliferating cell nuclear antigen (PCNA) is a DNA clamp that acts as a processivity factor for DNA polymerase δ in eukaryotic cells and is essential for replication. PCNA is a homotrimer and achieves its processivity by encircling the DNA, where it acts as a scaffold to recruit proteins involved in DNA replication, DNA repair, chromatin remodeling and epigenetics. Many proteins interact with PCNA via the two known PCNA-interacting motifs PCNA-interacting peptide (PIP) box and AlkB homologue 2 PCNA interacting motif (APIM). Proteins binding to PCNA via the PIP-box are mainly involved in DNA replication whereas proteins binding to PCNA via APIM are mainly important in the context of genotoxic stress. Function The protein encoded by this gene is found in the nucleus and is a cofactor of DNA polymerase delta. The encoded protein acts as a homotrimer and helps increase the processivity of leading strand synthesis during DNA replication. In response to DNA damage, this protein i ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Homologous Recombination
Homologous recombination is a type of genetic recombination in which genetic information is exchanged between two similar or identical molecules of double-stranded or single-stranded nucleic acids (usually DNA as in cellular organisms but may be also RNA in viruses). Homologous recombination is widely used by cells to accurately DNA repair harmful breaks that occur on both strands of DNA, known as double-strand breaks (DSB), in a process called homologous recombinational repair (HRR). Homologous recombination also produces new combinations of DNA sequences during meiosis, the process by which eukaryotes make gamete cells, like sperm and egg cells in animals. These new combinations of DNA represent genetic variation in offspring, which in turn enables populations to adapt during the course of evolution. Homologous recombination is also used in horizontal gene transfer to exchange genetic material between different strains and species of bacteria and viruses. Horizon ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
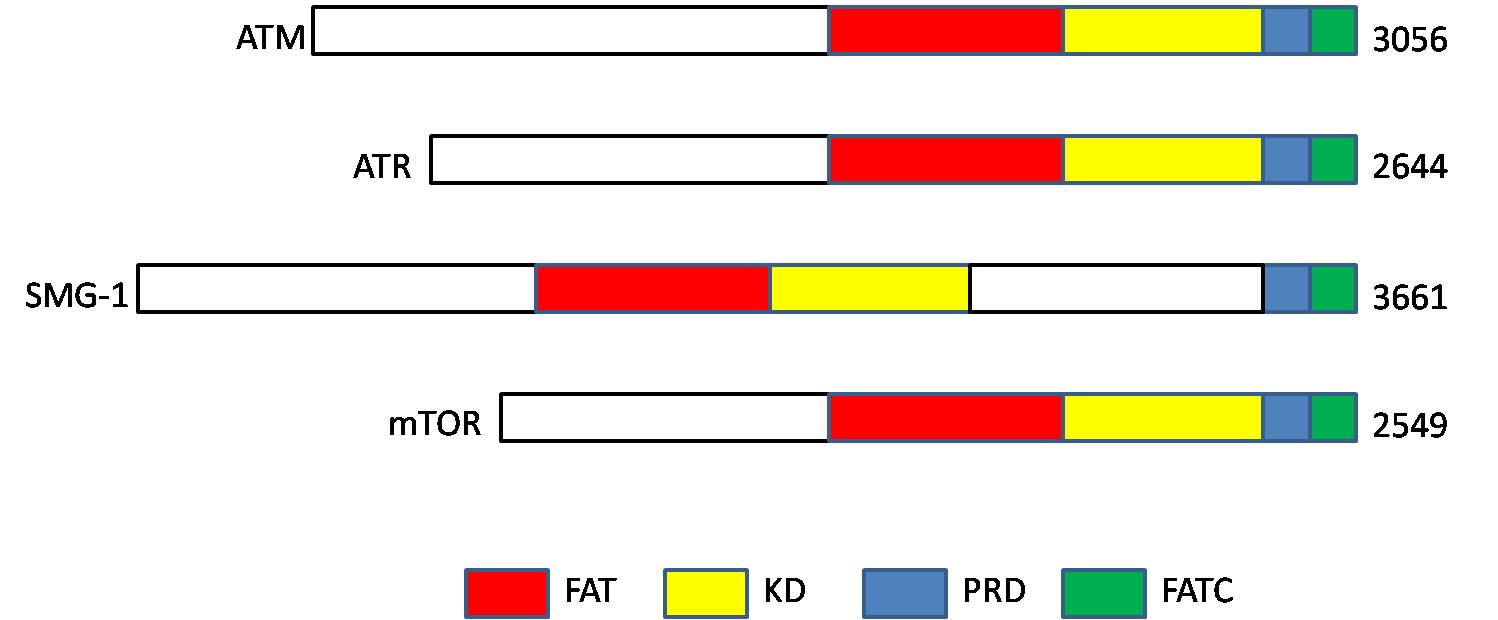
ATM Kinase
ATM serine/threonine kinase or Ataxia-telangiectasia mutated, symbol ATM, is a serine/ threonine protein kinase that is recruited and activated by DNA double-strand breaks. It phosphorylates several key proteins that initiate activation of the DNA damage checkpoint, leading to cell cycle arrest, DNA repair or apoptosis. Several of these targets, including p53, CHK2, BRCA1, NBS1 and H2AX are tumor suppressors. In 1995, the gene was discovered by Yosef Shiloh who named its product ATM since he found that its mutations are responsible for the disorder ataxia–telangiectasia. In 1998, the Shiloh and Kastan laboratories independently showed that ATM is a protein kinase whose activity is enhanced by DNA damage. Introduction Throughout the cell cycle DNA is monitored for damage. Damages result from errors during replication, by-products of metabolism, general toxic drugs or ionizing radiation. The cell cycle has different DNA damage checkpoints, which inhibit the next or maint ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
ATR Kinase
ATR may refer to: Medicine * Acute transfusion reaction * Ataxia telangiectasia and Rad3 related, a protein involved in DNA damage repair Science and mathematics * Advanced Test Reactor, nuclear research reactor at the Idaho National Laboratory, US * Attenuated total reflectance in infrared spectroscopy * Advanced tongue root, a phonological feature in linguistics * Atractyloside, a toxin and inhibitor of "ADP/ATP translocase" * ATR0, an axiom system in reverse mathematics Technology * Answer to reset, a message output by a contact Smart Card * Automatic target recognition, recognition ability * Autothermal reforming, a natural gas reforming technology Transport * ATR (aircraft manufacturer) an Italian-French aircraft manufacturer ** ATR 42 airliner ** ATR 72 airliner * IATA code for Atar International Airport *Andaman Trunk Road * Air Transport Rack, standards for plug-in electronic modules in aviation and elsewhere; various suppliers e.g. ARINC * Atmore (Amtrak station), Amt ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DNA Repair
DNA repair is a collection of processes by which a cell identifies and corrects damage to the DNA molecules that encode its genome. In human cells, both normal metabolic activities and environmental factors such as radiation can cause DNA damage, resulting in tens of thousands of individual molecular lesions per cell per day. Many of these lesions cause structural damage to the DNA molecule and can alter or eliminate the cell's ability to transcribe the gene that the affected DNA encodes. Other lesions induce potentially harmful mutations in the cell's genome, which affect the survival of its daughter cells after it undergoes mitosis. As a consequence, the DNA repair process is constantly active as it responds to damage in the DNA structure. When normal repair processes fail, and when cellular apoptosis does not occur, irreparable DNA damage may occur, including double-strand breaks and DNA crosslinkages (interstrand crosslinks or ICLs). This can eventually lead to malign ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
PRC2
PRC2 (polycomb repressive complex 2) is one of the two classes of polycomb-group proteins or (PcG). The other component of this group of proteins is PRC1 ( Polycomb Repressive Complex 1). This complex has histone methyltransferase activity and primarily methylates histone H3 on lysine 27 (i.e. H3K27me3), a mark of transcriptionally silent chromatin. PRC2 is required for initial targeting of genomic region (PRC Response Elements or PRE) to be silenced, while PRC1 is required for stabilizing this silencing and underlies cellular memory of silenced region after cellular differentiation. PRC1 also mono-ubiquitinates histone H2A on lysine 119 (H2AK119Ub1). These proteins are required for long term epigenetic silencing of chromatin and have an important role in stem cell differentiation and early embryonic development. PRC2 are present in most multicellular organisms. The mouse PRC2 has four subunits: Suz12 (zinc finger), Eed, Ezh1 or Ezh2 (SET domain with histone methyltransfe ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Semiconservative Replication
Semiconservative replication describe the mechanism of DNA replication in all known cells. DNA replication occurs on multiple origins of replication along the DNA template strand (antinsense strand). As the DNA double helix is unwound by helicase, replication occurs separately on each template strand (sense strand) in antiparallel directions. This process is known as semi-conservative replication because two copies of the original DNA molecule are produced, each copy conserving (replicating) the information from one half of the original DNA molecule. Each copy contains one original strand and one newly-synthesized strand. (Both copies should be identical, but this is not entirely assured.) The structure of DNA (as deciphered by James D. Watson and Francis Crick in 1953) suggested that each strand of the double helix would serve as a template for synthesis of a new strand. It was not known how newly synthesized strands combined with template strands to form two double helical DNA m ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
SLBP
Histone RNA hairpin-binding protein or stem-loop binding protein (SLBP) is a protein that in humans is encoded by the ''SLBP'' gene. Species distribution SLBP has been cloned from humans, '' C. elegans'', ''D. melanogaster'', '' X. laevis'', and sea urchins. The full length human protein has 270 amino acids (31 kDa) with a centrally located RNA binding domain (RBD). The 75 amino acid RBD is well conserved across species, however the remainder of SLBP is highly divergent in most organisms and not homologous to any other protein in the eukaryotic genomes. Function This gene encodes a protein that binds to the histone 3' UTR stem-loop structure in replication-dependent histone mRNAs. Histone mRNAs do not contain introns or polyadenylation signals, and are processed by a single endonucleolytic cleavage event downstream of the stem-loop. The stem-loop structure is essential for efficient processing of the histone pre-mRNA but this structure also controls the transport, tra ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Stem-loop
Stem-loop intramolecular base pairing is a pattern that can occur in single-stranded RNA. The structure is also known as a hairpin or hairpin loop. It occurs when two regions of the same strand, usually complementary in nucleotide sequence when read in opposite directions, base-pair to form a double helix that ends in an unpaired loop. The resulting structure is a key building block of many RNA secondary structures. As an important secondary structure of RNA, it can direct RNA folding, protect structural stability for messenger RNA (mRNA), provide recognition sites for RNA binding proteins, and serve as a substrate for enzymatic reactions. Formation and stability The formation of a stem-loop structure is dependent on the stability of the resulting helix and loop regions. The first prerequisite is the presence of a sequence that can fold back on itself to form a paired double helix. The stability of this helix is determined by its length, the number of mismatches or bulges it ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Polyadenylation
Polyadenylation is the addition of a poly(A) tail to an RNA transcript, typically a messenger RNA (mRNA). The poly(A) tail consists of multiple adenosine monophosphates; in other words, it is a stretch of RNA that has only adenine bases. In eukaryotes, polyadenylation is part of the process that produces mature mRNA for translation. In many bacteria, the poly(A) tail promotes degradation of the mRNA. It, therefore, forms part of the larger process of gene expression. The process of polyadenylation begins as the transcription of a gene terminates. The 3′-most segment of the newly made pre-mRNA is first cleaved off by a set of proteins; these proteins then synthesize the poly(A) tail at the RNA's 3′ end. In some genes these proteins add a poly(A) tail at one of several possible sites. Therefore, polyadenylation can produce more than one transcript from a single gene (alternative polyadenylation), similar to alternative splicing. The poly(A) tail is important for the nucl ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |