|
Secretomics
Secretomics is a type of proteomics which involves the analysis of the secretome—all the secreted proteins of a cell, tissue or organism. Secreted proteins are involved in a variety of physiological processes, including cell signaling and matrix remodeling, but are also integral to invasion and metastasis of malignant cells. Secretomics has thus been especially important in the discovery of biomarkers for cancer and understanding molecular basis of pathogenesis. The analysis of the insoluble fraction of the secretome (the extracellular matrix) has been termed matrisomics. History of the secretome In 2000 Tjalsma et al. coined the term 'secretome' in their study of the eubacterium ''B. subtilis''. They defined the secretome as all of the secreted proteins and secretory machinery of the bacteria. Using a database of protein sequences in ''B. subtilis'' and an algorithm that looked at cleavage sites and amino-terminal signal peptides characteristic of secreted proteins they were able ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Proteomics
Proteomics is the large-scale study of proteins. Proteins are vital parts of living organisms, with many functions such as the formation of structural fibers of muscle tissue, enzymatic digestion of food, or synthesis and replication of DNA. In addition, other kinds of proteins include antibodies that protect an organism from infection, and hormones that send important signals throughout the body. The proteome is the entire set of proteins produced or modified by an organism or system. Proteomics enables the identification of ever-increasing numbers of proteins. This varies with time and distinct requirements, or stresses, that a cell or organism undergoes. Proteomics is an interdisciplinary domain that has benefited greatly from the genetic information of various genome projects, including the Human Genome Project. It covers the exploration of proteomes from the overall level of protein composition, structure, and activity, and is an important component of functional genomi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Secretome
The secretome is the set of proteins expressed by an organism and secreted into the extracellular space. In humans, this subset of the proteome encompasses 13-20% of all proteins, including cytokines, growth factors, extracellular matrix proteins and regulators, and shed receptors. The secretome of a specific tissue can be measured by mass spectrometry and its analysis constitutes a type of proteomics known as secretomics. Definition The term ''secretome'' was coined by Tjalsma and colleagues in 2004 to denote all the factors secreted by a cell, along with the secretory pathway constituents. In 2010, this definition of secretome was revised to include only proteins secreted into the extracellular space. Related concepts include the matrisome, which is the subset of the secretome that includes extracellular matrix proteins and their associated proteins; the receptome, which includes all membrane receptors, and the adhesome, which includes all proteins involved in cell adhesion. ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Secretory Protein
A secretory protein is any protein, whether it be endocrine or exocrine, which is secreted by a cell. Secretory proteins include many hormones, enzymes, toxins, and antimicrobial peptides. Secretory proteins are synthesized in the endoplasmic reticulum. Production The production of a secretory protein starts like any other protein. The mRNA is produced and transported to the cytosol where it interacts with a free cytosolic ribosome. The part that is produced first, the N-terminal, contains a signal sequence consisting of 6 to 12 amino acids with hydrophobic side chains. This sequence is recognised by a cytosolic protein, SRP (Signal Recognition Particle), which stops the translation and aids in the transport of the mRNA-ribosome complex to an SRP receptor found in the membrane of the endoplasmic reticulum. When it arrives at the ER, the signal sequence is transferred to the translocon, a protein-conducting channel in the membrane that allows the newly synthesized polypeptide to ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Isoform
A protein isoform, or "protein variant", is a member of a set of highly similar proteins that originate from a single gene or gene family and are the result of genetic differences. While many perform the same or similar biological roles, some isoforms have unique functions. A set of protein isoforms may be formed from alternative splicings, variable promoter usage, or other post-transcriptional modifications of a single gene; post-translational modifications are generally not considered. (For that, see Proteoforms.) Through RNA splicing mechanisms, mRNA has the ability to select different protein-coding segments (exons) of a gene, or even different parts of exons from RNA to form different mRNA sequences. Each unique sequence produces a specific form of a protein. The discovery of isoforms could explain the discrepancy between the small number of protein coding regions genes revealed by the human genome project and the large diversity of proteins seen in an organism: different p ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Antibody
An antibody (Ab), also known as an immunoglobulin (Ig), is a large, Y-shaped protein used by the immune system to identify and neutralize foreign objects such as pathogenic bacteria and viruses. The antibody recognizes a unique molecule of the pathogen, called an antigen. Each tip of the "Y" of an antibody contains a paratope (analogous to a lock) that is specific for one particular epitope (analogous to a key) on an antigen, allowing these two structures to bind together with precision. Using this binding mechanism, an antibody can ''tag'' a microbe or an infected cell for attack by other parts of the immune system, or can neutralize it directly (for example, by blocking a part of a virus that is essential for its invasion). To allow the immune system to recognize millions of different antigens, the antigen-binding sites at both tips of the antibody come in an equally wide variety. In contrast, the remainder of the antibody is relatively constant. It only occurs in a few vari ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Chondrocyte
Chondrocytes (, from Greek χόνδρος, ''chondros'' = cartilage + κύτος, ''kytos'' = cell) are the only cells found in healthy cartilage. They produce and maintain the cartilaginous matrix, which consists mainly of collagen and proteoglycans. Although the word ''chondroblast'' is commonly used to describe an immature chondrocyte, the term is imprecise, since the progenitor of chondrocytes (which are mesenchymal stem cells) can differentiate into various cell types, including osteoblasts. Development From least- to terminally-differentiated, the chondrocytic lineage is: # Colony-forming unit-fibroblast # Mesenchymal stem cell / marrow stromal cell # Chondrocyte # Hypertrophic chondrocyte Mesenchymal ( mesoderm origin) stem cells are undifferentiated, meaning they can differentiate into a variety of generative cells commonly known as osteochondrogenic (or osteogenic, chondrogenic, osteoprogenitor, etc.) cells. When referring to bone, or in this case cartilage, the ori ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Stable Isotope Labeling By Amino Acids In Cell Culture
Stable Isotope Labeling by/with Amino acids in Cell culture (SILAC) is a technique based on mass spectrometry that detects differences in protein abundance among samples using non-radioactive isotopic labeling. It is a popular method for quantitative proteomics. Procedure Two populations of cells are cultivated in cell culture. One of the cell populations is fed with growth medium containing normal amino acids. In contrast, the second population is fed with growth medium containing amino acids labeled with stable (non-radioactive) heavy isotopes. For example, the medium can contain arginine labeled with six carbon-13 atoms (13C) instead of the normal carbon-12 (12C). When the cells are growing in this medium, they incorporate the heavy arginine into all of their proteins. Thereafter, all peptides containing a single arginine are 6 Da heavier than their normal counterparts. Alternatively, uniform labeling with 13C or 15N can be used. Proteins from both cell populations are combin ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Peptide-mass Fingerprint
In bio-informatics, a peptide-mass fingerprint or peptide-mass map is a mass spectrum of a mixture of peptides that comes from a digested protein being analyzed. The mass spectrum serves as a fingerprint in the sense that it is a pattern that can serve to identify the protein. The method for forming a peptide-mass fingerprint, developed in 1993, consists of isolating a protein, breaking it down into individual peptides, and determining the masses of the peptides through some form of mass spectrometry. Once formed, a peptide-mass fingerprint can be used to search in databases for related protein or even genomic sequences, making it a powerful tool for annotation of protein-coding genes. One major advantage to mass fingerprinting is that it is significantly faster to carry out than peptide sequencing, yet the results are equally useful. Disadvantages include the need for a single protein for analysis and the requirement that the protein sequence is located, at least with significant ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Chromatography
In chemical analysis, chromatography is a laboratory technique for the separation of a mixture into its components. The mixture is dissolved in a fluid solvent (gas or liquid) called the ''mobile phase'', which carries it through a system (a column, a capillary tube, a plate, or a sheet) on which a material called the ''stationary phase'' is fixed. Because the different constituents of the mixture tend to have different affinities for the stationary phase and are retained for different lengths of time depending on their interactions with its surface sites, the constituents travel at different apparent velocities in the mobile fluid, causing them to separate. The separation is based on the differential partitioning between the mobile and the stationary phases. Subtle differences in a compound's partition coefficient result in differential retention on the stationary phase and thus affect the separation. Chromatography may be preparative or analytical. The purpose of preparati ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protease
A protease (also called a peptidase, proteinase, or proteolytic enzyme) is an enzyme that catalyzes (increases reaction rate or "speeds up") proteolysis, breaking down proteins into smaller polypeptides or single amino acids, and spurring the formation of new protein products. They do this by cleaving the peptide bonds within proteins by hydrolysis, a reaction where water breaks bonds. Proteases are involved in many biological functions, including digestion of ingested proteins, protein catabolism (breakdown of old proteins), and cell signaling. In the absence of functional accelerants, proteolysis would be very slow, taking hundreds of years. Proteases can be found in all forms of life and viruses. They have independently evolved multiple times, and different classes of protease can perform the same reaction by completely different catalytic mechanisms. Hierarchy of proteases Based on catalytic residue Proteases can be classified into seven broad groups: * Serine protea ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Complementary DNA
In genetics, complementary DNA (cDNA) is DNA synthesized from a single-stranded RNA (e.g., messenger RNA (mRNA) or microRNA (miRNA)) template in a reaction catalyzed by the enzyme reverse transcriptase. cDNA is often used to express a specific protein in a cell that does not normally express that protein (i.e., heterologous expression), or to sequence or quantify mRNA molecules using DNA based methods (qPCR, RNA-seq). cDNA that codes for a specific protein can be transferred to a recipient cell for expression, often bacterial or yeast expression systems. cDNA is also generated to analyze transcriptomic profiles in bulk tissue, single cells, or single nuclei in assays such as microarrays, qPCR, and RNA-seq. cDNA is also produced naturally by retroviruses (such as HIV-1, HIV-2, simian immunodeficiency virus, etc.) and then integrated into the host's genome, where it creates a provirus. The term ''cDNA'' is also used, typically in a bioinformatics context, to refer to ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
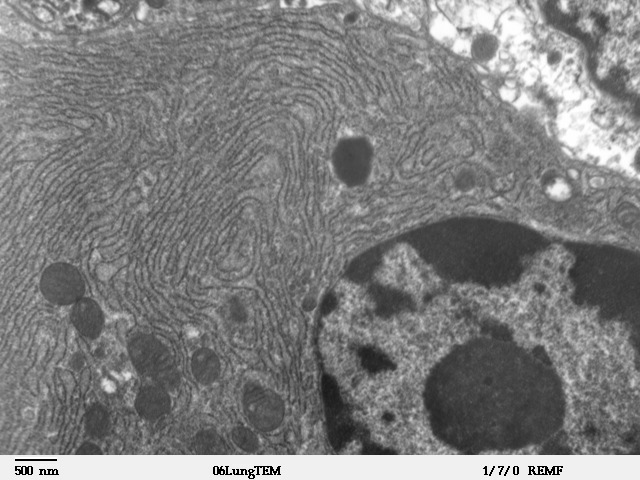
Endoplasmic Reticulum
The endoplasmic reticulum (ER) is, in essence, the transportation system of the eukaryotic cell, and has many other important functions such as protein folding. It is a type of organelle made up of two subunits – rough endoplasmic reticulum (RER), and smooth endoplasmic reticulum (SER). The endoplasmic reticulum is found in most eukaryotic cells and forms an interconnected network of flattened, membrane-enclosed sacs known as cisternae (in the RER), and tubular structures in the SER. The membranes of the ER are continuous with the outer nuclear membrane. The endoplasmic reticulum is not found in red blood cells, or spermatozoa. The two types of ER share many of the same proteins and engage in certain common activities such as the synthesis of certain lipids and cholesterol. Different types of cells contain different ratios of the two types of ER depending on the activities of the cell. RER is found mainly toward the nucleus of cell and SER towards the cell membrane or plasma ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
.jpg)