|
SMK Box Riboswitch
The SMKbox riboswitch (also known as SAM-III) is an RNA element that regulates gene expression in bacteria. The SMK box riboswitch is found in the 5' UTR of the MetK gene in lactic acid bacteria. The structure of this element changes upon binding to S-adenosyl methionine (SAM) to a conformation that blocks the shine-dalgarno sequence and blocks translation of the gene. There are other known SAM-binding riboswitches such as SAM-I and SAM-II, but these appear to share no similarity in sequence or structure to SAM-III. Structure The crystal structure of the riboswitch from ''E. faecalis'' was solved by X-ray crystallography. The structure showed that the most conserved nucleotides involved in SAM binding were organised around a junction between three helices. In some species there are large insertions of up to 210 nucleotides within this structure. See also * SAH riboswitch * SAM-I riboswitch * SAM-II riboswitch * SAM-IV riboswitch SAM-IV riboswitches are a kind of ri ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Nucleic Acid Secondary Structure
Nucleic acid secondary structure is the basepairing interactions within a single nucleic acid polymer or between two polymers. It can be represented as a list of bases which are paired in a nucleic acid molecule. The secondary structures of biological DNAs and RNAs tend to be different: biological DNA mostly exists as fully base paired double helices, while biological RNA is single stranded and often forms complex and intricate base-pairing interactions due to its increased ability to form hydrogen bonds stemming from the extra hydroxyl group in the ribose sugar. In a non-biological context, secondary structure is a vital consideration in the nucleic acid design of nucleic acid structures for DNA nanotechnology and DNA computing, since the pattern of basepairing ultimately determines the overall structure of the molecules. Fundamental concepts Base pairing In molecular biology, two nucleotides on opposite complementary DNA or RNA strands that are connected via hydrogen b ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
X-ray Crystallography
X-ray crystallography is the experimental science determining the atomic and molecular structure of a crystal, in which the crystalline structure causes a beam of incident X-rays to diffract into many specific directions. By measuring the angles and intensities of these diffracted beams, a crystallographer can produce a three-dimensional picture of the density of electrons within the crystal. From this electron density, the mean positions of the atoms in the crystal can be determined, as well as their chemical bonds, their crystallographic disorder, and various other information. Since many materials can form crystals—such as salts, metals, minerals, semiconductors, as well as various inorganic, organic, and biological molecules—X-ray crystallography has been fundamental in the development of many scientific fields. In its first decades of use, this method determined the size of atoms, the lengths and types of chemical bonds, and the atomic-scale differences among various mat ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
SAM–SAH Riboswitch
The SAM–SAH riboswitch is a conserved RNA structure in certain bacteria that binds ''S''-adenosylmethionine (SAM) and ''S''-adenosylhomocysteine (SAH) and is therefore presumed to be a riboswitch. SAM–SAH riboswitches do not share any apparent structural resemblance to known riboswitches that bind SAM or SAH. The binding affinities for both compounds are similar, but binding for SAH is at least somewhat stronger. SAM–SAH riboswitches are exclusively found in Rhodobacterales, an order of alphaproteobacteria. They are always found in the apparent 5' untranslated regions of ''metK'' genes, which encode the enzyme ( Methionine adenosyltransferase) that synthesizes SAM. Given this gene association, it was proposed that SAM–SAH riboswitches more likely function as SAM-sensing RNAs. SAM–SAH riboswitches are relatively small among known riboswitches, which might relate to their inability to discriminate against SAH. However, the ability to reject SAH as a ligand might not ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
SAM-Chlorobi RNA Motif
The SAM-Chlorobi RNA motif is a conserved RNA structure that was identified by bioinformatics. The RNAs are found only in bacteria classified as within the phylum Chlorobiota. These RNAs are always in the 5' untranslated regions of operons that contain ''metK'' and ''ahcY'' genes. ''metK'' genes encode methionine adenosyltransferase, which synthesizes S-adenosyl methionine (SAM), and ''ahcY'' genes encode S-adenosylhomocysteine hydrolase, which degrade the related metabolite S-Adenosyl-L-homocysteine (SAH). In fact all predicted ''metK'' and ''ahcY'' genes within Chlorobiota bacteria as of 2010 are preceded by predicted SAM-Chlorobi RNAs. Predicted promoter sequences are consistently found upstream of SAM-Chlorobi RNAs, and these promoter sequences imply that SAM-Chlorobi RNAs are indeed transcribed as RNAs. The promoter sequences are commonly associated with strong transcription in the phyla Chlorobiota and Bacteroidota The phylum Bacteroidota (synonym Bacteroidete ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
SAM-VI Riboswitch
SAM-VI is a member of the riboswitch family. It is predominantly found in ''Bifidobacterium'' and exhibits some similarities to the SAM-III ( Smk box) riboswitch class, but lacks most of the highly conserved nucleotides of SAM-III class. SAM-VI aptamers bind the cofactor S-adenosylmethinine SAM (a key metabolite in sulphur metabolism) and discriminate strongly against S-adenosylhomocysteine SAH. The class was discovered by further analysis of Bifido-''meK'' motif RNAs. See also * SAM-I riboswitch * SAM-II riboswitch * SAM-III riboswitch * SAM-IV riboswitch SAM-IV riboswitches are a kind of riboswitch that specifically binds S-adenosylmethionine (SAM), a cofactor used in many methylation reactions. Originally identified by bioinformatics, SAM-IV riboswitches are largely confined to the Actinomyce ... * SAM-V riboswitch References {{reflist Cis-regulatory RNA elements Riboswitch ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
SAM-V Riboswitch
SAM-V riboswitch is the fifth known riboswitch to bind S-adenosyl methionine (SAM). It was first discovered in the marine bacterium '' Candidatus Pelagibacter ubique'' and can also be found in marine metagenomes. SAM-V features a similar consensus sequence and secondary structure as the binding site of SAM-II riboswitch, but bioinformatics scans cluster the two aptamers independently. These similar binding pockets suggest that the two riboswitches have undergone convergent evolution. SAM-binding was confirmed using equilibrium dialysis. The riboswitch has been characterised as a 'tandem riboswitch' - it is able to regulate both translation and transcription. When SAM is present in high concentration, SAM-II will bind its ligand and form a terminator stem to halt transcription. If SAM exists in lower concentrations, SAM-V will be transcribed and, if SAM concentration should then increase, it can bind SAM and occlude the Shine-Dalgarno sequence of the downstream open reading fr ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
SAM-IV Riboswitch
SAM-IV riboswitches are a kind of riboswitch that specifically binds S-adenosylmethionine (SAM), a cofactor used in many methylation reactions. Originally identified by bioinformatics, SAM-IV riboswitches are largely confined to the Actinomycetales, an order of Bacteria. Conserved features of SAM-IV riboswitch and experiments imply that they probably share a similar SAM-binding site to another class of SAM-binding riboswitches called SAM-I riboswitches. However, the scaffolds of these two types of riboswitch appear to be quite distinct. The structural relationship between these riboswitch types has been studied. See also * SAM-I riboswitch * SAM-II riboswitch The SAM-II riboswitch is a RNA element found predominantly in Alphaproteobacteria that binds S-adenosyl methionine (SAM). Its structure and sequence appear to be unrelated to the SAM riboswitch found in Gram-positive bacteria. This SAM riboswit ... * SAM-III riboswitch * SAM-V riboswitch * SAM-VI riboswitch R ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
SAM Riboswitch (S-box Leader)
The SAM riboswitch (also known as the S-box leader and the SAM-I riboswitch) is found upstream of a number of genes which code for proteins involved in methionine or cysteine biosynthesis in Gram-positive bacteria. Two SAM riboswitches in ''Bacillus subtilis'' that were experimentally studied act at the level of transcription termination control. The predicted secondary structure consists of a complex stem-loop region followed by a single stem-loop terminator region. An alternative and mutually exclusive form involves bases in the 3' segment of helix 1 with those in the 5' region of helix 5 to form a structure termed the anti-terminator form. When SAM is unbound, the anti-terminator sequence sequesters the terminator sequence so the terminator is unable to form, allowing the polymerase to read-through the downstream gene.Winkler, W., Nahvi, A., Sudarsan, N., Barrick, J., and Breaker, R. (2003) An mRNA structure that controls gene expression by binding S-adenosylmethionine. ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
SAH Riboswitch
SAH riboswitches are a kind of riboswitch that bind S-adenosylhomocysteine (SAH). When the coenzyme S-adenosylmethionine (SAM) is used in a methylation reaction, SAH is produced. SAH riboswitches typically up-regulate genes involved in recycling SAH to create more SAM (or the metabolically related methionine). This is particularly relevant to cells, because high levels of SAH can be toxic. Originally identified by bioinformatics, SAH riboswitches are apparent in many species of bacteria, predominantly certain Pseudomonadota Pseudomonadota (synonym Proteobacteria) is a major phylum of Gram-negative bacteria. The renaming of phyla in 2021 remains controversial among microbiologists, many of whom continue to use the earlier names of long standing in the literature. The ... and Actinomycetota. The atomic-resolution 3-dimensional structure of an SAH riboswitch has been solved using X-ray crystallography. References External links * Cis-regulatory RNA elements Riboswi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
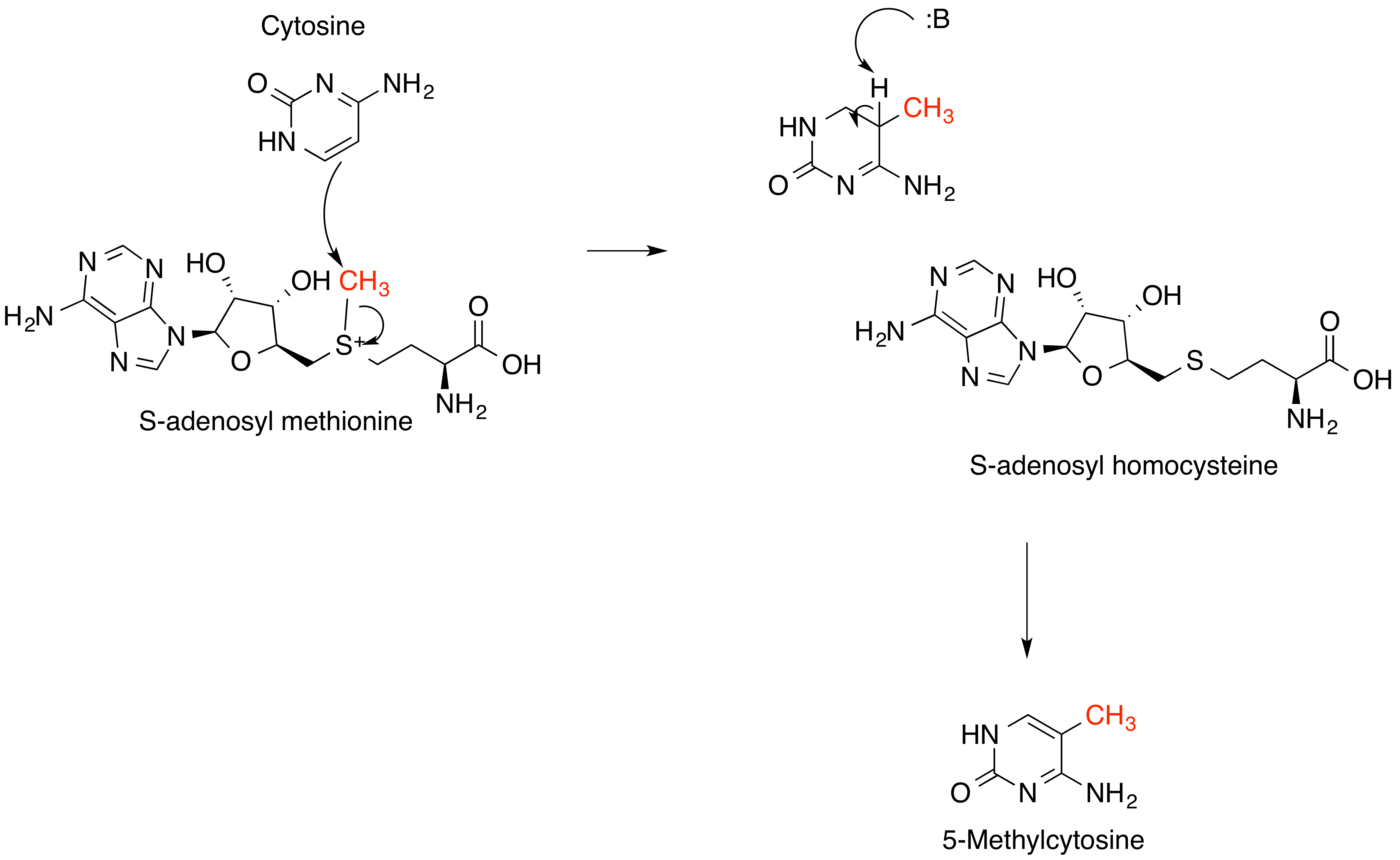
S-adenosyl Methionine
''S''-Adenosyl methionine (SAM), also known under the commercial names of SAMe, SAM-e, or AdoMet, is a common cosubstrate involved in methyl group transfers, transsulfuration, and aminopropylation. Although these anabolic reactions occur throughout the body, most SAM is produced and consumed in the liver. More than 40 methyl transfers from SAM are known, to various substrates such as nucleic acids, proteins, lipids and secondary metabolites. It is made from adenosine triphosphate (ATP) and methionine by methionine adenosyltransferase. SAM was first discovered by Giulio Cantoni in 1952. In bacteria, SAM is bound by the SAM riboswitch, which regulates genes involved in methionine or cysteine biosynthesis. In eukaryotic cells, SAM serves as a regulator of a variety of processes including DNA, tRNA, and rRNA methylation; immune response; amino acid metabolism; transsulfuration; and more. In plants, SAM is crucial to the biosynthesis of ethylene, an important plant hormone and sig ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
SAM-II Riboswitch
The SAM-II riboswitch is a RNA element found predominantly in Alphaproteobacteria that binds S-adenosyl methionine (SAM). Its structure and sequence appear to be unrelated to the SAM riboswitch found in Gram-positive bacteria. This SAM riboswitch is located upstream of the metA and metC genes in Agrobacterium tumefaciens, and other methionine and SAM biosynthesis genes in other alpha-proteobacteria. Like the other SAM riboswitch, it probably functions to turn off expression of these genes in response to elevated SAM levels. A significant variant of SAM-II riboswitches was found in ''Pelagibacter ubique'' and related marine bacteria and called SAM-V. Also, like many structured RNAs, SAM-II riboswitches can tolerate long loops between their stems. Structure The SAM-II riboswitch is short with less than 70 nucleotides and is structurally relatively simple being composed of a single hairpin A hairpin or hair pin is a long device used to hold a person's hair in place. It ma ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Riboswitch
In molecular biology, a riboswitch is a regulatory segment of a messenger RNA molecule that binds a small molecule, resulting in a change in production of the proteins encoded by the mRNA. Thus, an mRNA that contains a riboswitch is directly involved in regulating its own activity, in response to the concentrations of its effector molecule. The discovery that modern organisms use RNA to bind small molecules, and discriminate against closely related analogs, expanded the known natural capabilities of RNA beyond its ability to code for proteins, catalyze reactions, or to bind other RNA or protein macromolecules. The original definition of the term "riboswitch" specified that they directly sense small-molecule metabolite concentrations. Although this definition remains in common use, some biologists have used a broader definition that includes other cis-regulatory RNAs. However, this article will discuss only metabolite-binding riboswitches. Most known riboswitches occur in bac ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |