|
S100A1
Protein S100-A1, also known as S100 calcium-binding protein A1 is a protein which in humans is encoded by the ''S100A1'' gene. S100A1 is highly expressed in cardiac and skeletal muscle, and localizes to Z-discs and sarcoplasmic reticulum. S100A1 has shown promise as an effective candidate for gene therapy to treat post- myocardially infarcted cardiac tissue. Structure S100A1 is a member of the S100 family of proteins expressed in cardiac muscle, skeletal muscle and brain, with highest density at Z-lines and sarcoplasmic reticulum. S100A1 contains 4 EF-hand calcium-binding motifs in its dimerized form, and can exist as either a hetero or homodimer. The S100A1 homodimer is high affinity (nanomolar range or tighter), and is formed through hydrophobic packing of an X-type 4-helix bundle created between helices 1, 1', 4, and 4'. Protein nuclear magnetic resonance spectroscopy structural information on the homodimeric form of this protein shows that each monomer is helical and cont ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
S-100 Protein
The S100 proteins are a family of low molecular-weight proteins found in vertebrates characterized by two calcium-binding sites that have helix-loop-helix ("EF-hand-type") conformation. At least 21 different S100 proteins are known. They are encoded by a family of genes whose symbols use the ''S100'' prefix, for example, ''S100A1'', ''S100A2'', ''S100A3''. They are also considered as damage-associated molecular pattern molecules (DAMPs), and knockdown of aryl hydrocarbon receptor downregulates the expression of S100 proteins in THP-1 cells. Structure Most S100 proteins consist of two identical polypeptides (homodimeric), which are held together by noncovalent bonds. They are structurally similar to calmodulin. They differ from calmodulin, though, on the other features. For instance, their expression pattern is cell-specific, i.e. they are expressed in particular cell types. Their expression depends on environmental factors. In contrast, calmodulin is a ubiquitous and univer ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
EF-hand
The EF hand is a helix–loop–helix structural domain or ''motif'' found in a large family of calcium-binding proteins. The EF-hand motif contains a helix–loop–helix topology, much like the spread thumb and forefinger of the human hand, in which the Ca2+ ions are coordinated by ligands within the loop. The motif takes its name from traditional nomenclature used in describing the protein parvalbumin, which contains three such motifs and is probably involved in muscle relaxation via its calcium-binding activity. The EF-hand consists of two alpha helices linked by a short loop region (usually about 12 amino acids) that usually binds calcium ions. EF-hands also appear in each structural domain of the signaling protein calmodulin and in the muscle protein troponin-C. Calcium ion binding site The calcium ion is coordinated in a pentagonal bipyramidal configuration. The six residues involved in the binding are in positions 1, 3, 5, 7, 9 and 12; these residues are denoted by ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
EF Hand
The EF hand is a helix–loop–helix structural domain or ''motif'' found in a large family of calcium-binding proteins. The EF-hand motif contains a helix–loop–helix topology, much like the spread thumb and forefinger of the human hand, in which the Ca2+ ions are coordinated by ligands within the loop. The motif takes its name from traditional nomenclature used in describing the protein parvalbumin, which contains three such motifs and is probably involved in muscle relaxation via its calcium-binding activity. The EF-hand consists of two alpha helices linked by a short loop region (usually about 12 amino acids) that usually binds calcium ions. EF-hands also appear in each structural domain of the signaling protein calmodulin and in the muscle protein troponin-C. Calcium ion binding site The calcium ion is coordinated in a pentagonal bipyramidal configuration. The six residues involved in the binding are in positions 1, 3, 5, 7, 9 and 12; these residues are denoted by X ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Alpha Helix
The alpha helix (α-helix) is a common motif in the secondary structure of proteins and is a right hand-helix conformation in which every backbone N−H group hydrogen bonds to the backbone C=O group of the amino acid located four residues earlier along the protein sequence. The alpha helix is also called a classic Pauling–Corey–Branson α-helix. The name 3.613-helix is also used for this type of helix, denoting the average number of residues per helical turn, with 13 atoms being involved in the ring formed by the hydrogen bond. Among types of local structure in proteins, the α-helix is the most extreme and the most predictable from sequence, as well as the most prevalent. Discovery In the early 1930s, William Astbury showed that there were drastic changes in the X-ray fiber diffraction of moist wool or hair fibers upon significant stretching. The data suggested that the unstretched fibers had a coiled molecular structure with a characteristic repeat of ≈. Astb ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cysteine
Cysteine (symbol Cys or C; ) is a semiessential proteinogenic amino acid with the formula . The thiol side chain in cysteine often participates in enzymatic reactions as a nucleophile. When present as a deprotonated catalytic residue, sometimes the symbol Cyz is used. The deprotonated form can generally be described by the symbol Cym as well. The thiol is susceptible to oxidation to give the disulfide derivative cystine, which serves an important structural role in many proteins. In this case, the symbol Cyx is sometimes used. When used as a food additive, it has the E number E920. Cysteine is encoded by the codons UGU and UGC. The sulfur-containing amino acids cysteine and methionine are more easily oxidized than the other amino acids. Structure Like other amino acids (not as a residue of a protein), cysteine exists as a zwitterion. Cysteine has chirality in the older / notation based on homology to - and -glyceraldehyde. In the newer ''R''/''S'' system of designating chi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
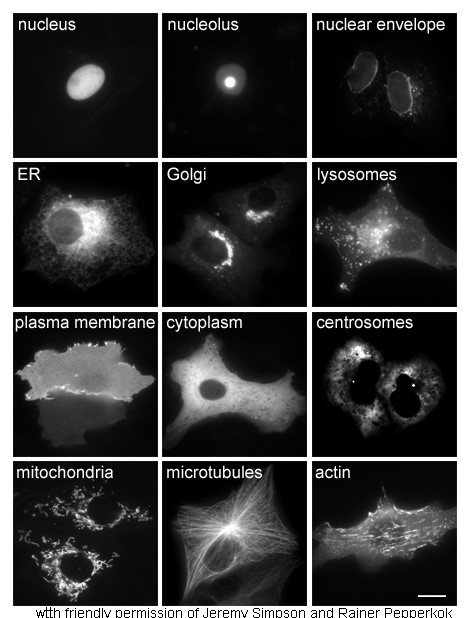
Cytoplasm
In cell biology, the cytoplasm is all of the material within a eukaryotic cell, enclosed by the cell membrane, except for the cell nucleus. The material inside the nucleus and contained within the nuclear membrane is termed the nucleoplasm. The main components of the cytoplasm are cytosol (a gel-like substance), the organelles (the cell's internal sub-structures), and various cytoplasmic inclusions. The cytoplasm is about 80% water and is usually colorless. The submicroscopic ground cell substance or cytoplasmic matrix which remains after exclusion of the cell organelles and particles is groundplasm. It is the hyaloplasm of light microscopy, a highly complex, polyphasic system in which all resolvable cytoplasmic elements are suspended, including the larger organelles such as the ribosomes, mitochondria, the plant plastids, lipid droplets, and vacuoles. Most cellular activities take place within the cytoplasm, such as many metabolic pathways including glycolysis, and proces ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cell Nucleus
The cell nucleus (pl. nuclei; from Latin or , meaning ''kernel'' or ''seed'') is a membrane-bound organelle found in eukaryotic cells. Eukaryotic cells usually have a single nucleus, but a few cell types, such as mammalian red blood cells, have no nuclei, and a few others including osteoclasts have many. The main structures making up the nucleus are the nuclear envelope, a double membrane that encloses the entire organelle and isolates its contents from the cellular cytoplasm; and the nuclear matrix, a network within the nucleus that adds mechanical support. The cell nucleus contains nearly all of the cell's genome. Nuclear DNA is often organized into multiple chromosomes – long stands of DNA dotted with various proteins, such as histones, that protect and organize the DNA. The genes within these chromosomes are structured in such a way to promote cell function. The nucleus maintains the integrity of genes and controls the activities of the cell by regulating gene expres ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein
Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residues. Proteins perform a vast array of functions within organisms, including catalysing metabolic reactions, DNA replication, responding to stimuli, providing structure to cells and organisms, and transporting molecules from one location to another. Proteins differ from one another primarily in their sequence of amino acids, which is dictated by the nucleotide sequence of their genes, and which usually results in protein folding into a specific 3D structure that determines its activity. A linear chain of amino acid residues is called a polypeptide. A protein contains at least one long polypeptide. Short polypeptides, containing less than 20–30 residues, are rarely considered to be proteins and are commonly called peptides. The individual amino acid residues are bonded together by peptide bonds and adjacent amino acid residues. The sequence of amino acid residue ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Dissociation Constant
In chemistry, biochemistry, and pharmacology, a dissociation constant (K_D) is a specific type of equilibrium constant that measures the propensity of a larger object to separate (dissociate) reversibly into smaller components, as when a complex falls apart into its component molecules, or when a salt splits up into its component ions. The dissociation constant is the inverse of the association constant. In the special case of salts, the dissociation constant can also be called an ionization constant. For a general reaction: : A_\mathit B_\mathit \mathit A + \mathit B in which a complex \ce_x \ce_y breaks down into ''x'' A subunits and ''y'' B subunits, the dissociation constant is defined as : K_D = \frac where and ''x'' B''y''are the equilibrium concentrations of A, B, and the complex A''x'' B''y'', respectively. One reason for the popularity of the dissociation constant in biochemistry and pharmacology is that in the frequently encount ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cell Cycle
The cell cycle, or cell-division cycle, is the series of events that take place in a cell that cause it to divide into two daughter cells. These events include the duplication of its DNA (DNA replication) and some of its organelles, and subsequently the partitioning of its cytoplasm, chromosomes and other components into two daughter cells in a process called cell division. In cells with nuclei ( eukaryotes, i.e., animal, plant, fungal, and protist cells), the cell cycle is divided into two main stages: interphase and the mitotic (M) phase (including mitosis and cytokinesis). During interphase, the cell grows, accumulating nutrients needed for mitosis, and replicates its DNA and some of its organelles. During the mitotic phase, the replicated chromosomes, organelles, and cytoplasm separate into two new daughter cells. To ensure the proper replication of cellular components and division, there are control mechanisms known as cell cycle checkpoints after each of the key steps ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cellular Differentiation
Cellular differentiation is the process in which a stem cell alters from one type to a differentiated one. Usually, the cell changes to a more specialized type. Differentiation happens multiple times during the development of a multicellular organism as it changes from a simple zygote to a complex system of tissues and cell types. Differentiation continues in adulthood as adult stem cells divide and create fully differentiated daughter cells during tissue repair and during normal cell turnover. Some differentiation occurs in response to antigen exposure. Differentiation dramatically changes a cell's size, shape, membrane potential, metabolic activity, and responsiveness to signals. These changes are largely due to highly controlled modifications in gene expression and are the study of epigenetics. With a few exceptions, cellular differentiation almost never involves a change in the DNA sequence itself. Although metabolic composition does get altered quite dramaticall ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |