|
Sirohydrochlorin Cobaltochelatase
The enzyme sirohydrochlorin cobaltochelatase (EC 4.99.1.3) catalyzes the reaction :cobalt-sirohydrochlorin + 2 H+ = sirohydrochlorin + Co2+ In the forward direction of reactions towards cobalamin in anaerobic bacteria, the two substrates of this enzyme are sirohydrochlorin and Co2+; its two products are cobalt-sirohydrochlorin and H+. This enzyme belongs to the family of lyases, specifically the "catch-all" class of lyases that do not fit into any other sub-class. The systematic name of this enzyme class is cobalt-sirohydrochlorin cobalt-lyase (sirohydrochlorin-forming). Other names in common use include CbiK, CbiX, CbiXS, anaerobic cobalt chelatase, cobaltochelatase mbiguous'', and sirohydrochlorin cobalt-lyase (incorrect). This enzyme is part of the biosynthetic pathway to cobalamin (vitamin B12) in bacteria such as ''Salmonella typhimurium'' and ''Bacillus megaterium''. It has also been identified as the enzyme which inserts nickel into sirohydrochlorin in the biosynthes ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Catalysis
Catalysis () is the process of increasing the rate of a chemical reaction by adding a substance known as a catalyst (). Catalysts are not consumed in the reaction and remain unchanged after it. If the reaction is rapid and the catalyst recycles quickly, very small amounts of catalyst often suffice; mixing, surface area, and temperature are important factors in reaction rate. Catalysts generally react with one or more reactants to form intermediates that subsequently give the final reaction product, in the process of regenerating the catalyst. Catalysis may be classified as either homogeneous, whose components are dispersed in the same phase (usually gaseous or liquid) as the reactant, or heterogeneous, whose components are not in the same phase. Enzymes and other biocatalysts are often considered as a third category. Catalysis is ubiquitous in chemical industry of all kinds. Estimates are that 90% of all commercially produced chemical products involve catalysts at some s ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Lyase
In biochemistry, a lyase is an enzyme that catalyzes the breaking (an elimination reaction) of various chemical bonds by means other than hydrolysis (a substitution reaction) and oxidation, often forming a new double bond or a new ring structure. The reverse reaction is also possible (called a Michael reaction). For example, an enzyme that catalyzed this reaction would be a lyase: : ATP → cAMP + PPi Lyases differ from other enzymes in that they require only one substrate for the reaction in one direction, but two substrates for the reverse reaction. Nomenclature Systematic names are formed as "''substrate group-lyase''." Common names include decarboxylase, dehydratase, aldolase, etc. When the product is more important, synthase may be used in the name, e.g. phosphosulfolactate synthase (EC 4.4.1.19, Michael addition of sulfite to phosphoenolpyruvate). A combination of both an elimination and a Michael addition is seen in O-succinylhomoserine (thiol)-lyase (MetY or MetZ) which ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Data Bank
The Protein Data Bank (PDB) is a database for the three-dimensional structural data of large biological molecules, such as proteins and nucleic acids. The data, typically obtained by X-ray crystallography, NMR spectroscopy, or, increasingly, cryo-electron microscopy, and submitted by biologists and biochemists from around the world, are freely accessible on the Internet via the websites of its member organisations (PDBe, PDBj, RCSB, and BMRB). The PDB is overseen by an organization called the Worldwide Protein Data Bank, wwPDB. The PDB is a key in areas of structural biology, such as structural genomics. Most major scientific journals and some funding agencies now require scientists to submit their structure data to the PDB. Many other databases use protein structures deposited in the PDB. For example, SCOP and CATH classify protein structures, while PDBsum provides a graphic overview of PDB entries using information from other sources, such as Gene ontology. History Two force ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Tertiary Structure
Protein tertiary structure is the three dimensional shape of a protein. The tertiary structure will have a single polypeptide chain "backbone" with one or more protein secondary structures, the protein domains. Amino acid side chains may interact and bond in a number of ways. The interactions and bonds of side chains within a particular protein determine its tertiary structure. The protein tertiary structure is defined by its atomic coordinates. These coordinates may refer either to a protein domain or to the entire tertiary structure.Branden C. and Tooze J. "Introduction to Protein Structure" Garland Publishing, New York. 1990 and 1991. A number of tertiary structures may fold into a quaternary structure.Kyte, J. "Structure in Protein Chemistry." Garland Publishing, New York. 1995. History The science of the tertiary structure of proteins has progressed from one of hypothesis to one of detailed definition. Although Emil Fischer had suggested proteins were made of polypept ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cobalamin Biosynthesis
Cobalamin biosynthesis is the process by which bacteria and archea make Vitamin B12, cobalamin, vitamin B12. Many steps are involved in converting aminolevulinic acid via uroporphyrinogen III and adenosylcobyric acid to the final forms in which it is used by enzymes in both the producing organisms and other species, including humans who acquire it through their diet. The feature which distinguishes the two main Biosynthesis, biosynthetic routes is whether the cobalt that is at the catalytic site in the coenzyme is incorporated early (in anaerobic organisms) or late (in aerobic organisms) and whether oxygen is required. In both cases, the macrocycle that will form a coordination complex with the cobalt ion is a corrin ring, specifically one with seven Carboxylic acid, carboxylate groups called cobyrinic acid. Subsequently, amide groups are formed on all but one of the carboxylates, giving cobyric acid, and the cobalt is ligand, ligated by an adenosine, adenosyl group. In the final p ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cofactor F430
F430 is the cofactor (sometimes called the coenzyme) of the enzyme methyl coenzyme M reductase (MCR). MCR catalyzes the reaction that releases methane in the final step of methanogenesis: : + HS–CoB → + CoB–S–S–CoM It is found only in methanogenic Archaea and anaerobic methanotrophic Archaea. It occurs in relatively high concentrations in archaea that are involved in reverse methanogenesis: these can contain up to 7% by weight of the nickel protein. Structure The trivial name cofactor F430 was assigned in 1978 based on the properties of a yellow sample extracted from ''Methanobacterium thermoautotrophicum'', which had a spectroscopic maximum at 430 nm. It was identified as the MCR cofactor in 1982 and the complete structure was deduced by X-ray crystallography and NMR spectroscopy. Coenzyme F430 features a reduced porphyrin in a macrocyclic ring system called a corphin. In addition, it possesses two additional rings in comparison to the standard tetrapyrrole ( ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
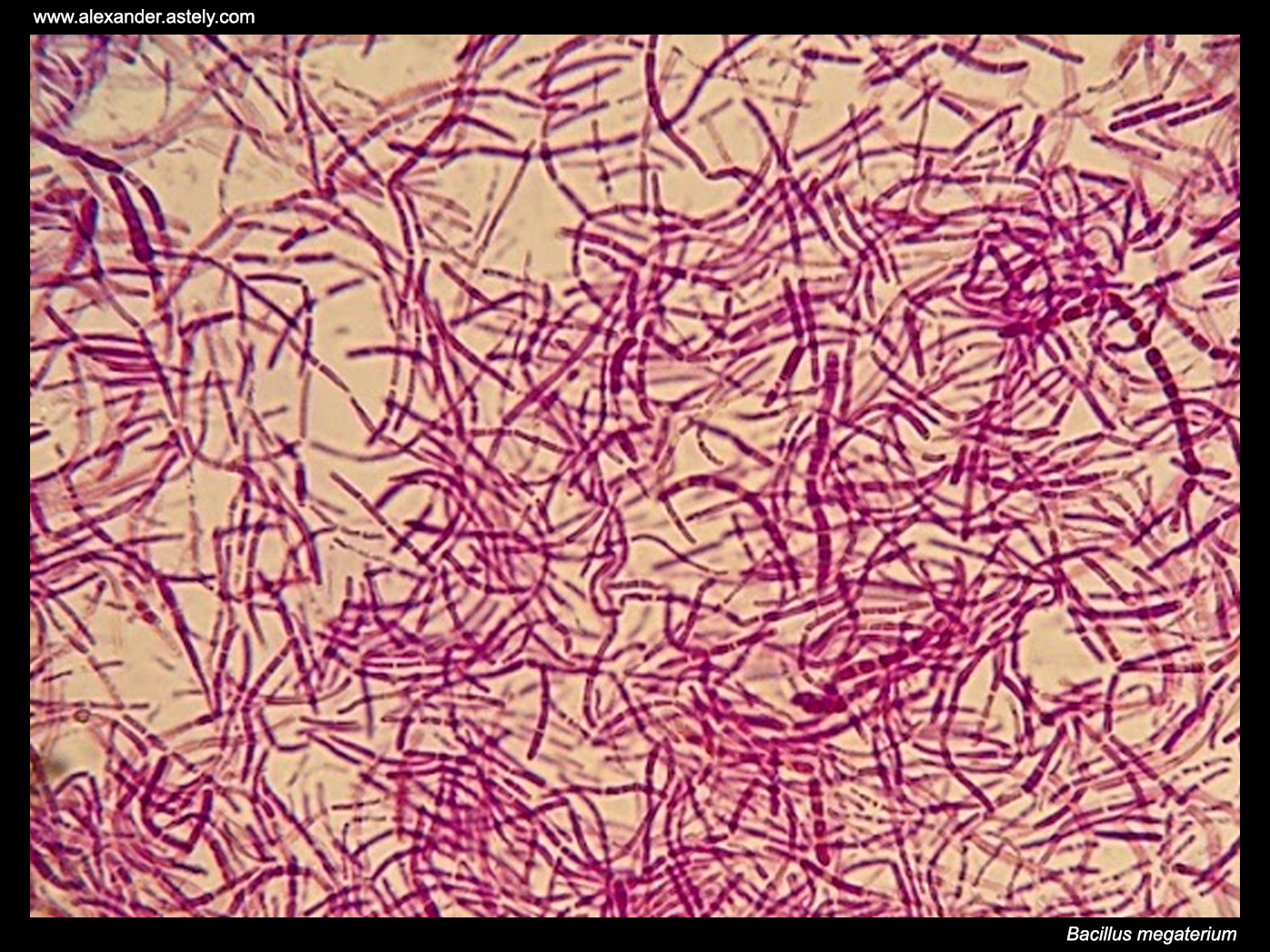
Bacillus Megaterium
''Bacillus megaterium'' is a rod-like, Gram-positive, mainly aerobic spore forming bacterium found in widely diverse habitats.De Vos, P. ''et al.'' Bergey's Manual of Systematic Bacteriology: Volume 3: The Firmicutes. ''Springer'' (2009) It has a cell length of up to 4 µm and a diameter of 1.5 µm, which is quite large for a bacteria. The cells often occur in pairs and chains, where the cells are joined together by polysaccharides on the cell walls. In the 1960s, prior to the utilization of ''Bacillus subtilis'' for this purpose, ''B. megaterium'' was the main model organism among Gram-positive bacteria for intensive studies on biochemistry, sporulation and bacteriophages. Recently, its popularity has started increasing in the field of biotechnology for its recombinant protein production capacity.Bunk, B. ''et al.'' A short story about a big magic bug. ''Bioengineered Bugs'' 1:85–91 (2010) This species has been recently transferred into the genus ''Priestia''. The c ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Salmonella Typhimurium
''Salmonella enterica'' subsp. ''enterica'' is a subspecies of ''Salmonella enterica'', the rod-shaped, flagellated, aerobic, Gram-negative bacterium. Many of the pathogenic serovars of the ''S. enterica'' species are in this subspecies, including that responsible for typhoid. Serovars ''S. enterica'' subsp. ''enterica'' contains a large number of serovars which can infect a broad range of vertebrate hosts. The individual members range from being highly host-adapted (only able to infect a narrow range of species) to displaying a broad host range. A number of techniques are currently used to differentiate between serotypes. These include looking for the presence or absence of antigens, phage typing, molecular fingerprinting and biotyping, where serovars are differentiated by which nutrients they are able to ferment. A possible factor in determining the host range of particular serovars is phage-mediated acquisition of a small number of genetic elements that enable infection of a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
List Of Enzymes
This article lists enzymes by their classification in the International Union of Biochemistry and Molecular Biology's Enzyme Commission (EC) numbering system. * List of EC numbers (EC 5) * List of EC numbers (EC 6) :Oxidoreductases (EC 1) (Oxidoreductase) *Dehydrogenase * Luciferase *DMSO reductase :EC 1.1 (act on the CH-OH group of donors) * :EC 1.1.1 (with NAD+ or NADP+ as acceptor) ** Alcohol dehydrogenase (NAD) ** Alcohol dehydrogenase (NADP) **Homoserine dehydrogenase ** Aminopropanol oxidoreductase **Diacetyl reductase **Glycerol dehydrogenase **Propanediol-phosphate dehydrogenase ** glycerol-3-phosphate dehydrogenase (NAD+) ** D-xylulose reductase **L-xylulose reductase **Lactate dehydrogenase **Malate dehydrogenase **Isocitrate dehydrogenase ** HMG-CoA reductase * :EC 1.1.2 (with a cytochrome as acceptor) * :EC 1.1.3 (with oxygen as acceptor) **Glucose oxidase **L-gulonolactone oxidase **Thiamine oxidase **Xanthine oxidase * :EC 1.1.4 (with a disul ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Hydrogen Ion
A hydrogen ion is created when a hydrogen atom loses or gains an electron. A positively charged hydrogen ion (or proton) can readily combine with other particles and therefore is only seen isolated when it is in a gaseous state or a nearly particle-free space. Due to its extremely high charge density of approximately 2×1010 times that of a sodium ion, the bare hydrogen ion cannot exist freely in solution as it readily hydrates, i.e., bonds quickly. The hydrogen ion is recommended by IUPAC as a general term for all ions of hydrogen and its isotopes. Depending on the charge of the ion, two different classes can be distinguished: positively charged ions and negatively charged ions. Cation (positively charged) A hydrogen atom is made up of a nucleus with charge +1, and a single electron. Therefore, the only positively charged ion possible has charge +1. It is noted H+. Depending on the isotope in question, the hydrogen cation has different names: * Hydron: general name referri ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Organocobalt
Organocobalt chemistry is the chemistry of organometallic compounds containing a carbon to cobalt chemical bond. Organocobalt compounds are involved in several organic reactions and the important biomolecule vitamin B12 has a cobalt-carbon bond. Many organocobalt compounds exhibit useful catalytic properties, the preeminent example being dicobalt octacarbonyl. Alkyl complexes Most fundamental are the cobalt complexes with only alkyl ligands. Examples include Co(4-norbornyl)4 and its cation. Alkylcobalt is represented by vitamin B12 and related enzymes. In methylcobalamin the ligand is a methyl group, which is electrophilic. in vitamin B12, the alkyl ligand is an adenosyl group. Related to vitamin B12 are cobalt porphyrins, dimethylglyoximates, and related complexes of Schiff base ligands. These synthetic compounds also form alkyl derivatives that undergo diverse reactions reminiscent of the biological processes. The weak cobalt(III)-carbon bond in vitamin B12 analogues c ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |