|
Ribosomopathy
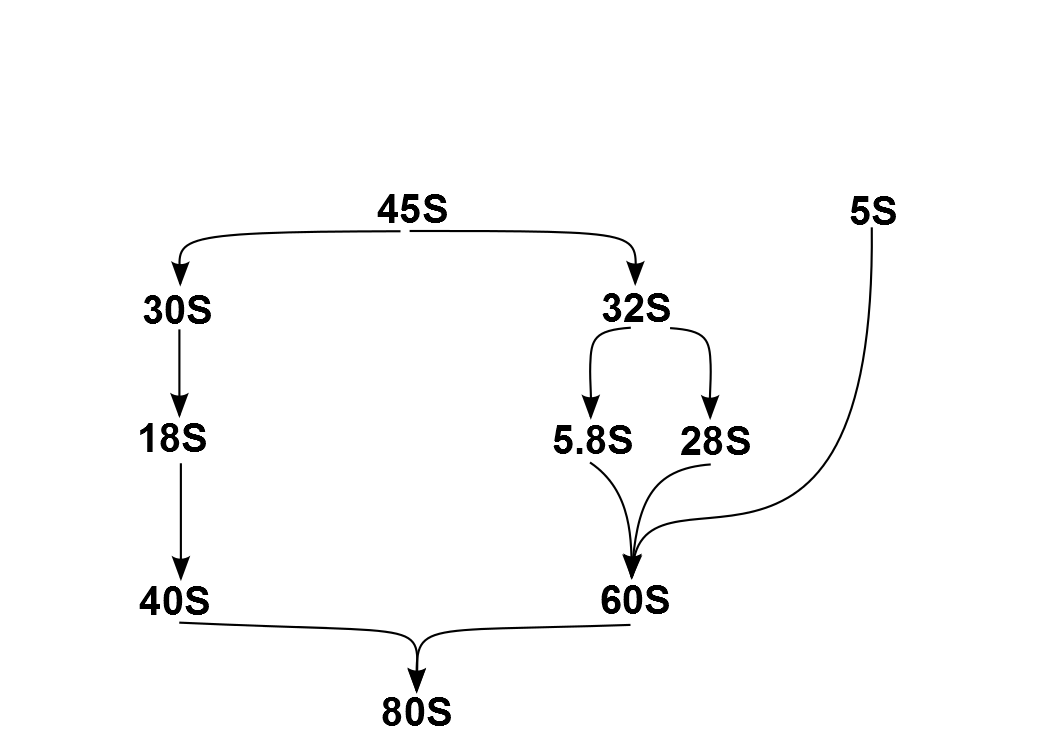
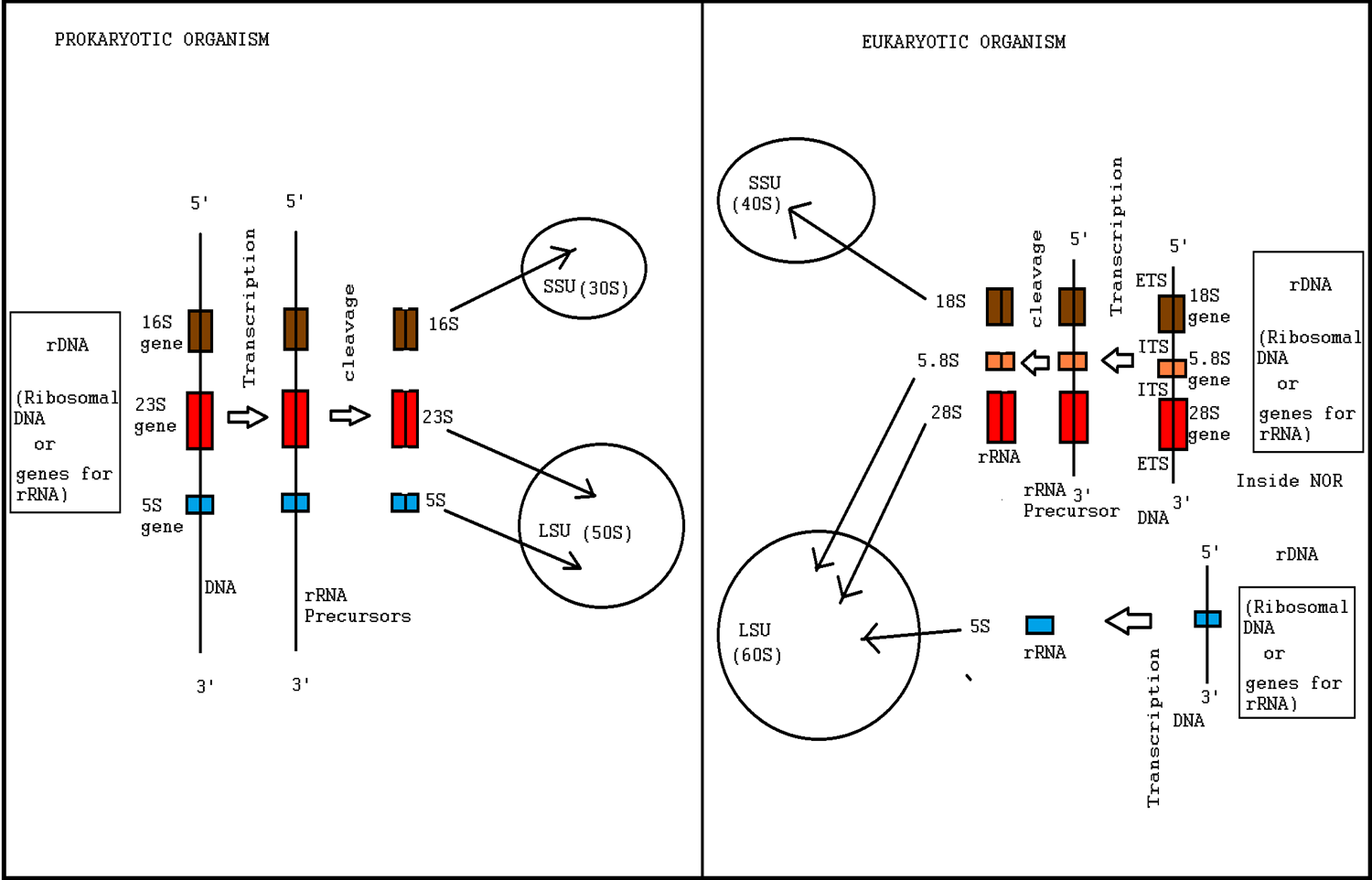
Ribosomopathies are diseases caused by abnormalities in the structure or function of ribosomal component proteins or rRNA genes, or other genes whose products are involved in ribosome biogenesis. Ribosomes Ribosomes are essential for protein synthesis in all living organisms. Prokaryotic and eukaryotic ribosomes both contain a scaffold of ribosomal RNA (rRNA) on which are arrayed an extensive variety of ribosomal proteins (RP). Ribosomopathies can arise from abnormalities of either rRNA or the various RPs. The nomenclature of rRNA subunits is derived from each component's Svedberg unit, which is an ultracentrifuge sedimentation coefficient, that is affected by mass and also shape. These S units of the rRNA subunits cannot simply be added because they represent measures of sedimentation rate rather than of mass. Eukaryotic ribosomes are somewhat larger and more complex than prokaryotic ribosomes. The overall 80S eukaryotic rRNA structure is composed of a large 60S subunit (LSU) ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
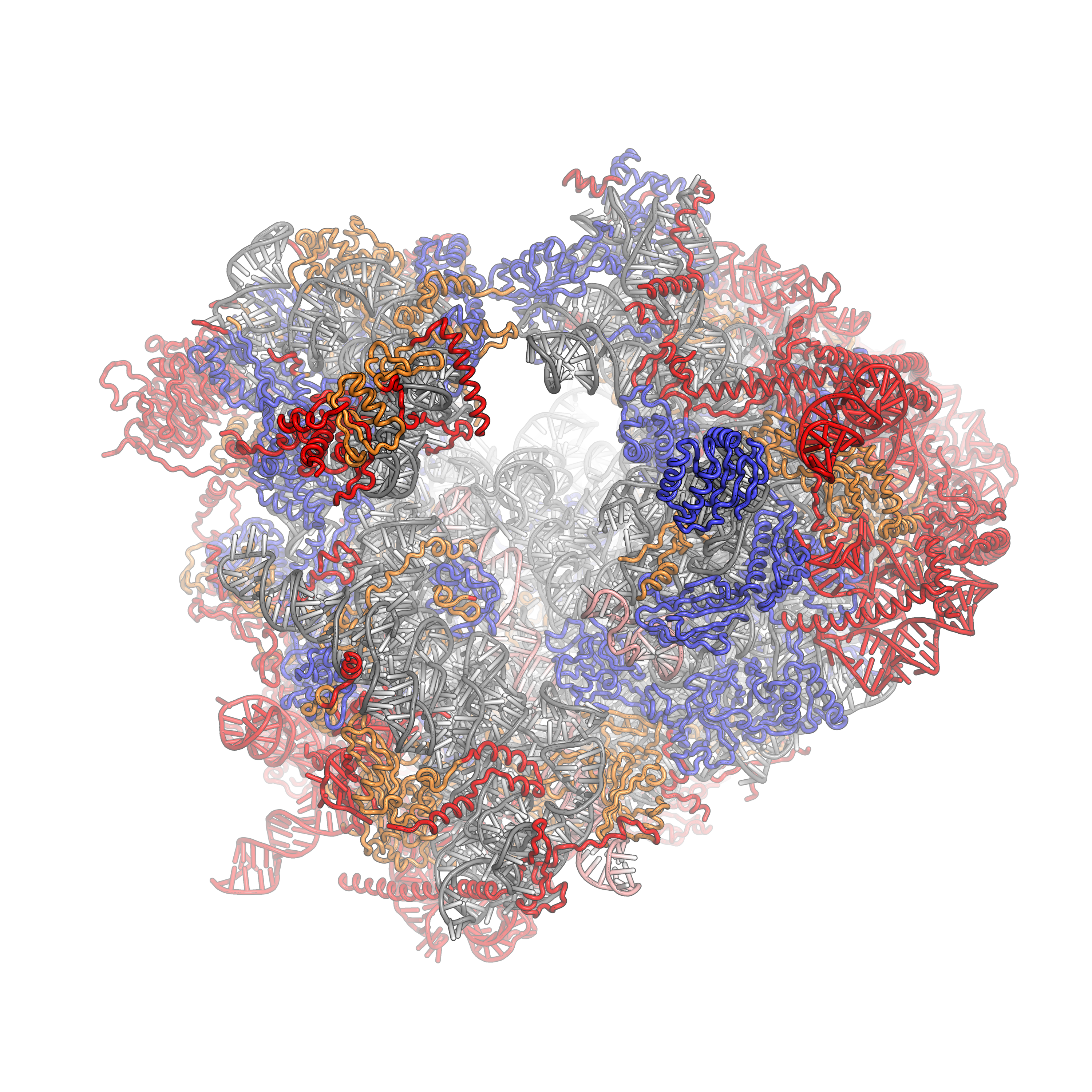
Eukaryotic Ribosome (80S)
Ribosomes are a large and complex molecular machine that catalyzes the synthesis of proteins, referred to as translation. The ribosome selects aminoacylated transfer RNAs (tRNAs) based on the sequence of a protein-encoding messenger RNA (mRNA) and covalently links the amino acids into a polypeptide chain. Ribosomes from all organisms share a highly conserved catalytic center. However, the ribosomes of eukaryotes (animals, plants, fungi, and large number unicellular organisms all with a nucleus) are much larger than prokaryotic (bacterial and archaeal) ribosomes and subject to more complex regulation and biogenesis pathways. Eukaryotic ribosomes are also known as 80S ribosomes, referring to their sedimentation coefficients in Svedberg units, because they sediment faster than the prokaryotic (70S) ribosomes. Eukaryotic ribosomes have two unequal subunits, designated small subunit (40S) and large subunit (60S) according to their sedimentation coefficients. Both subunits contain doze ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ribosome Biogenesis
Ribosome biogenesis is the process of making ribosomes. In prokaryotes, this process takes place in the cytoplasm with the transcription of many ribosome gene operons. In eukaryotes, it takes place both in the cytoplasm and in the nucleolus. It involves the coordinated function of over 200 proteins in the synthesis and processing of the three prokaryotic or four eukaryotic rRNAs, as well as assembly of those rRNAs with the ribosomal proteins. Most of the ribosomal proteins fall into various energy-consuming enzyme families including ATP-dependent RNA helicases, AAA-ATPases, GTPases, and kinases. About 60% of a cell's energy is spent on ribosome production and maintenance. Ribosome biogenesis is a very tightly regulated process, and it is closely linked to other cellular activities like growth and division. Some have speculated that in the origin of life, ribosome biogenesis predates cells, and that genes and cells evolved to enhance the reproductive capacity of ribosomes. R ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Eukaryotic Small Ribosomal Subunit (40S)
The eukaryotic small ribosomal subunit (40S) is the smaller subunit of the eukaryotic 80S ribosomes, with the other major component being the large ribosomal subunit (60S). The "40S" and "60S" names originate from the convention that ribosomal particles are denoted according to their sedimentation coefficients in Svedberg units. It is structurally and functionally related to the 30S subunit of 70S prokaryotic ribosomes. However, the 40S subunit is much larger than the prokaryotic 30S subunit and contains many additional protein segments, as well as rRNA expansion segments. Function The 40S subunit contains the decoding center which monitors the complementarity of tRNA and mRNA in protein translation. It is the largest component of several translation initiation complexes, including the 43S and 48S preinitiation complexes (PICs), being bound by several eukaryotic initiation factors, including eIF1, eIF1A, and eIF3. The 40S ribosomal subunit is also tightly bound by the HCV IRES ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
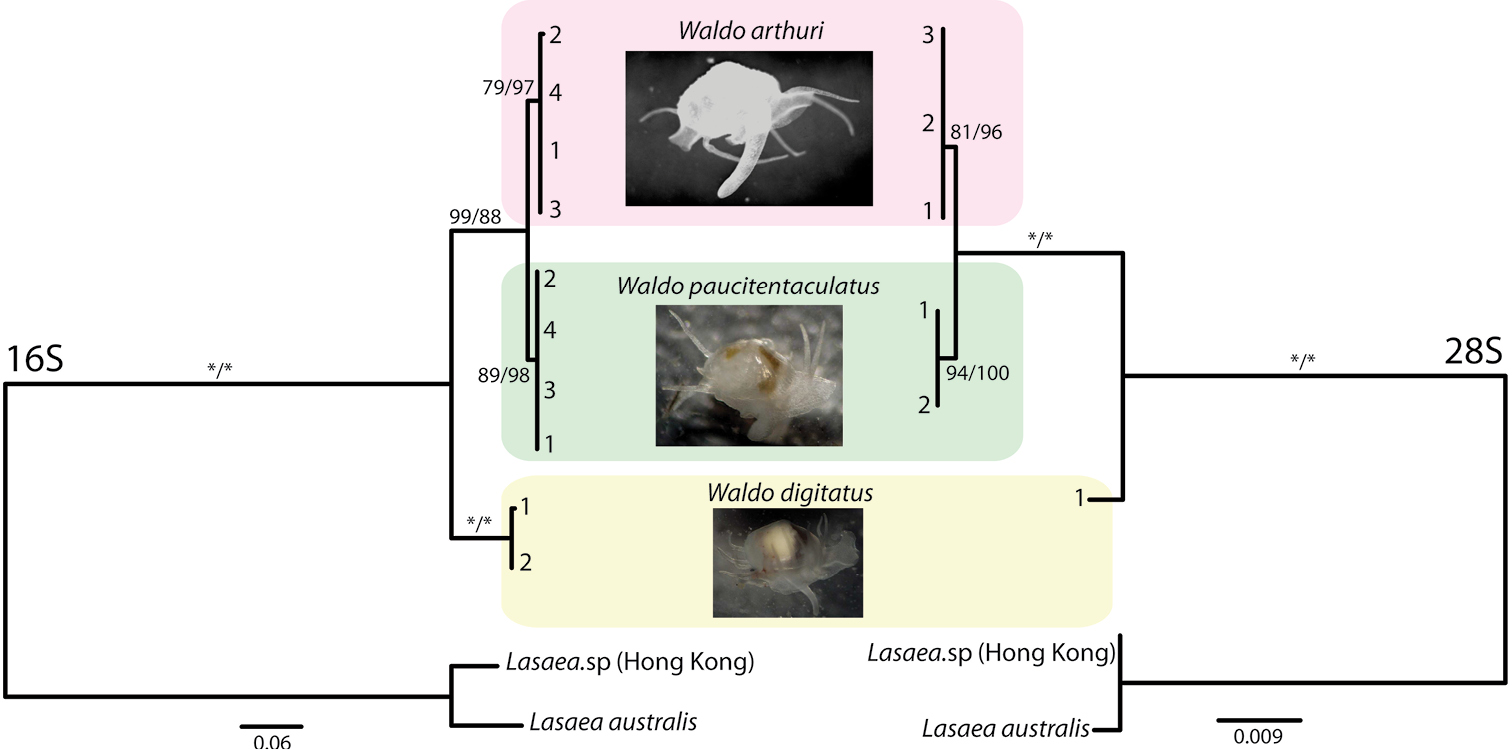
MT-RNR2
Mitochondrially encoded 16S RNA (often abbreviated as ''16S'') is the mitochondrial large subunit ribosomal RNA that in humans is encoded by the MT-RNR2 gene. The MT-RNR2 gene also encodes the Humanin polypeptide that has been the target of Alzheimer's disease research. The 16S rRNA is the mitochondrial homologue of the prokaryotic 23S and eukaryotic nuclear 28S ribosomal RNAs. See also * Mitochondrial DNA * Humanin Humanin is a micropeptide encoded in the mitochondrial genome by the 16S ribosomal RNA gene, MT-RNR2. Its structure contains a three-turn α-helix, and no symmetry. In ''in vitro'' and animal models, it appears to have cytoprotective effects. ... References Ribosomal RNA Human mitochondrial genes {{genetics-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
12S Ribosomal RNA
Mitochondrially encoded 12S ribosomal RNA (often abbreviated as 12S or 12S rRNA), also known as Mitochondrial-derived peptide MOTS-c or Mitochondrial open reading frame of the 12S rRNA-c is the SSU rRNA of the mitochondrial ribosome. In humans, 12S is encoded by the ''MT-RNR1'' gene and is 959 nucleotides long. MT-RNR1 is one of the 37 genes contained in animal mitochondria genomes. Their 2 rRNA, 22 tRNA and 13 mRNA genes are very useful in phylogenetic studies, in particular the 12S and 16S rRNAs. The 12S rRNA is the mitochondrial homologue of the prokaryotic 16S and eukaryotic nuclear 18S ribosomal RNAs. Mutations in the MT-RNR1 gene may be associated with hearing loss. Structure The ''MT-RNR1'' gene is located on the p arm of the mitochondrial DNA at position 12 and it spans 953 base pairs. Function The ''MT-RNR1'' gene encodes for a protein responsible for regulating insulin sensitivity and metabolic homeostasis. The protein acts as an inhibitor of the folate cycle ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cell Nucleus
The cell nucleus (pl. nuclei; from Latin or , meaning ''kernel'' or ''seed'') is a membrane-bound organelle found in eukaryotic cells. Eukaryotic cells usually have a single nucleus, but a few cell types, such as mammalian red blood cells, have no nuclei, and a few others including osteoclasts have many. The main structures making up the nucleus are the nuclear envelope, a double membrane that encloses the entire organelle and isolates its contents from the cellular cytoplasm; and the nuclear matrix, a network within the nucleus that adds mechanical support. The cell nucleus contains nearly all of the cell's genome. Nuclear DNA is often organized into multiple chromosomes – long stands of DNA dotted with various proteins, such as histones, that protect and organize the DNA. The genes within these chromosomes are structured in such a way to promote cell function. The nucleus maintains the integrity of genes and controls the activities of the cell by regulating gene expres ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
RNA Polymerase III
In eukaryote cells, RNA polymerase III (also called Pol III) is a protein that transcribes DNA to synthesize ribosomal 5S rRNA, tRNA and other small RNAs. The genes transcribed by RNA Pol III fall in the category of "housekeeping" genes whose expression is required in all cell types and most environmental conditions. Therefore, the regulation of Pol III transcription is primarily tied to the regulation of cell growth and the cell cycle, thus requiring fewer regulatory proteins than RNA polymerase II. Under stress conditions however, the protein Maf1 represses Pol III activity. Rapamycin is another Pol III inhibitor via its direct target TOR. Transcription The process of transcription (by any polymerase) involves three main stages: *Initiation, requiring construction of the RNA polymerase complex on the gene's promoter *Elongation, the synthesis of the RNA transcript *Termination, the finishing of RNA transcription and disassembly of the RNA polymerase complex Initiation In ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Pseudogene
Pseudogenes are nonfunctional segments of DNA that resemble functional genes. Most arise as superfluous copies of functional genes, either directly by DNA duplication or indirectly by Reverse transcriptase, reverse transcription of an mRNA transcript. Pseudogenes are usually identified when genome sequence analysis finds gene-like sequences that lack regulatory sequences needed for Transcription (biology), transcription or Translation (biology), translation, or whose coding sequences are obviously defective due to Frameshift mutation, frameshifts or premature stop codons. Most non-bacterial genomes contain many pseudogenes, often as many as functional genes. This is not surprising, since various biological processes are expected to accidentally create pseudogenes, and there are no specialized mechanisms to remove them from genomes. Eventually pseudogenes may be deleted from their genomes by chance DNA replication or DNA repair errors, or they may accumulate so many mutational cha ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Tandemly Arrayed Genes
Tandemly arrayed genes (TAGs) are a gene cluster created by tandem duplications, a process in which one gene is duplicated and the copy is found adjacent to the original. They serve to encode large numbers of genes at a time. TAGs represent a large proportion of genes in a genome, including between 14% to 17% of the human, mouse, and rat genomes. TAG clusters may have as few as two genes, with small clusters predominating, but may consist of hundreds of genes. An example are tandem clusters of rRNA encoding genes. These genes are transcribed faster than they would be if only a single copy of the gene was available. Additionally, a single RNA gene may not be able to provide enough RNA, but tandem repeats of the gene allow sufficient RNA to be produced. For example, cells in a human embryo contain between five and ten million ribosomes, and cell number doubles within 24 hours. In order to provide the necessary ribosomes, multiple RNA polymerases must consecutively transcribe multiple ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
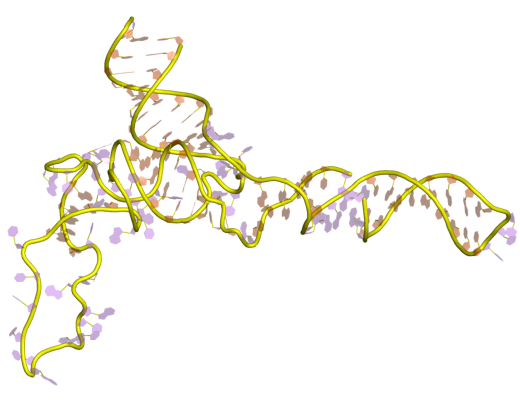
5S Ribosomal RNA
The 5S ribosomal RNA (5S rRNA) is an approximately 120 nucleotide-long ribosomal RNA molecule with a mass of 40 kDa. It is a structural and functional component of the large subunit of the ribosome in all domains of life (bacteria, archaea, and eukaryotes), with the exception of mitochondrial ribosomes of fungi and animals. The designation 5S refers to the molecule's sedimentation velocity in an ultracentrifuge, which is measured in Svedberg units (S). Biosynthesis In prokaryotes, the 5S rRNA gene is typically located in the rRNA operons downstream of the small and the large subunit rRNA, and co-transcribed into a polycistronic precursor. A particularity of eukaryotic nuclear genomes is the occurrence of multiple 5S rRNA gene copies (5S rDNA) clustered in tandem repeats, with copy number varying from species to species. Eukaryotic 5S rRNA is synthesized by RNA polymerase III, whereas other eukaryotic rRNAs are cleaved from a 45S precursor transcribed by RNA polymerase I. In ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
18S Ribosomal RNA
18S ribosomal RNA (abbreviated 18S rRNA) is a part of the ribosomal RNA. The S in 18S represents Svedberg units. 18S rRNA is an SSU rRNA, a component of the eukaryotic ribosomal small subunit (40S). 18S rRNA is the structural RNA for the small component of eukaryotic cytoplasmic ribosomes, and thus one of the basic components of all eukaryotic cells. 18S rRNA is the eukaryotic cytosolic homologue of 16S ribosomal RNA in prokaryotes and plastids. 18S rRNA is also a homologue of 12S ribosomal RNA in mitochondria. The genes coding for 18S rRNA are referred to as 18S rRNA genes. Sequence data from these genes is widely used in molecular analysis to reconstruct the evolutionary history of organisms, especially in vertebrates, as its slow evolutionary rate makes it suitable to reconstruct ancient divergences. Uses in phylogeny The small subunit (SSU) 18S rRNA gene is one of the most frequently used genes in phylogenetic studies and an important marker for random target polymerase ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
28S Ribosomal RNA
28S ribosomal RNA is the structural ribosomal RNA (rRNA) for the large subunit (LSU) of eukaryotic cytoplasmic ribosomes, and thus one of the basic components of all eukaryotic cells. It has a size of 25S in plants and 28S in mammals, hence the alias of 25S–28S rRNA. Combined with 5.8S rRNA to the 5' side, it is the eukaryotic nuclear homologue of the prokaryotic 23S and mitochondrial 16S ribosomal RNAs. Use in phylogeny The genes coding for 28S rRNA are referred to as 28S rDNA. The comparison of the sequences from these genes are sometimes used in molecular analysis to construct phylogenetic trees, for example in protists, fungi, insects, arachnids, tardigrades, and vertebrates. Structure The 28S rRNA is typically 4000–5000 nt long. Some eukaryotes cleave 28S rRNA into two parts before assembling both into the ribosome, a phenomenon termed the "hidden break". Databases Several databases provide alignments and annotations of LSU rRNA sequences for comparati ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |