|
Ribosomal Protein L13 Leader
L13 ribosomal protein leaders play a role in ribosome biogenesis as part of an autoregulatory mechanism to control the concentration of ribosomal proteins L13. Three structural classes of L13 ribosomal protein leaders were detected by different bioinformatics approaches: in ''B. subtilis'' and other low-GC Gram-positive bacteria., in ''E. coli'' and in Bacteroidia. Although these RNAs are expected to perform the same biological function, they do not appear to be structurally related to one another. The ''E. coli'' example has been experimentally confirmed, though the experiments are not comprehensive. The other two leader structures are thus far not based on experimental support. See also Ribosomal protein leader A ribosomal protein leader is a mechanism used in cells to control the cellular concentration of a protein that forms a part of the ribosome, and to make sure that the concentration is neither too high nor too low. Ribosomal protein leaders are RNA ... Reference ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Secondary Structure
Protein secondary structure is the three dimensional conformational isomerism, form of ''local segments'' of proteins. The two most common Protein structure#Secondary structure, secondary structural elements are alpha helix, alpha helices and beta sheets, though beta turns and omega loops occur as well. Secondary structure elements typically spontaneously form as an intermediate before the protein protein folding, folds into its three dimensional protein tertiary structure, tertiary structure. Secondary structure is formally defined by the pattern of hydrogen bonds between the Amine, amino hydrogen and carboxyl oxygen atoms in the peptide backbone chain, backbone. Secondary structure may alternatively be defined based on the regular pattern of backbone Dihedral angle#Dihedral angles of proteins, dihedral angles in a particular region of the Ramachandran plot regardless of whether it has the correct hydrogen bonds. The concept of secondary structure was first introduced by Kaj Ulrik ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Sequence Conservation
In evolutionary biology, conserved sequences are identical or similar sequences in nucleic acids ( DNA and RNA) or proteins across species ( orthologous sequences), or within a genome ( paralogous sequences), or between donor and receptor taxa ( xenologous sequences). Conservation indicates that a sequence has been maintained by natural selection. A highly conserved sequence is one that has remained relatively unchanged far back up the phylogenetic tree, and hence far back in geological time. Examples of highly conserved sequences include the RNA components of ribosomes present in all domains of life, the homeobox sequences widespread amongst Eukaryotes, and the tmRNA in Bacteria. The study of sequence conservation overlaps with the fields of genomics, proteomics, evolutionary biology, phylogenetics, bioinformatics and mathematics. History The discovery of the role of DNA in heredity, and observations by Frederick Sanger of variation between animal insulins in 1949, promp ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cis-regulatory Element
''Cis''-regulatory elements (CREs) or ''Cis''-regulatory modules (CRMs) are regions of non-coding DNA which regulate the transcription of neighboring genes. CREs are vital components of genetic regulatory networks, which in turn control morphogenesis, the development of anatomy, and other aspects of embryonic development, studied in evolutionary developmental biology. CREs are found in the vicinity of the genes that they regulate. CREs typically regulate gene transcription by binding to transcription factors. A single transcription factor may bind to many CREs, and hence control the expression of many genes ( pleiotropy). The Latin prefix ''cis'' means "on this side", i.e. on the same molecule of DNA as the gene(s) to be transcribed. CRMs are stretches of DNA, usually 100–1000 DNA base pairs in length, where a number of transcription factors can bind and regulate expression of nearby genes and regulate their transcription rates. They are labeled as ''cis'' because they are ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Attenuator (genetics)
In genetics, attenuation is a regulatory mechanism for some bacterial operons that results in premature termination of transcription. The canonical example of attenuation used in many introductory genetics textbooks, is ribosome-mediated attenuation of the ''trp'' operon. Ribosome-mediated attenuation of the ''trp'' operon relies on the fact that, in bacteria, transcription and translation proceed simultaneously. Attenuation involves a provisional stop signal (attenuator), located in the DNA segment that corresponds to the leader sequence of mRNA. During attenuation, the ribosome becomes stalled (delayed) in the attenuator region in the mRNA leader. Depending on the metabolic conditions, the attenuator either stops transcription at that point or allows read-through to the structural gene part of the mRNA and synthesis of the appropriate protein. Attenuation is a regulatory feature found throughout Archaea and Bacteria causing premature termination of transcription.Merino E, Yanof ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
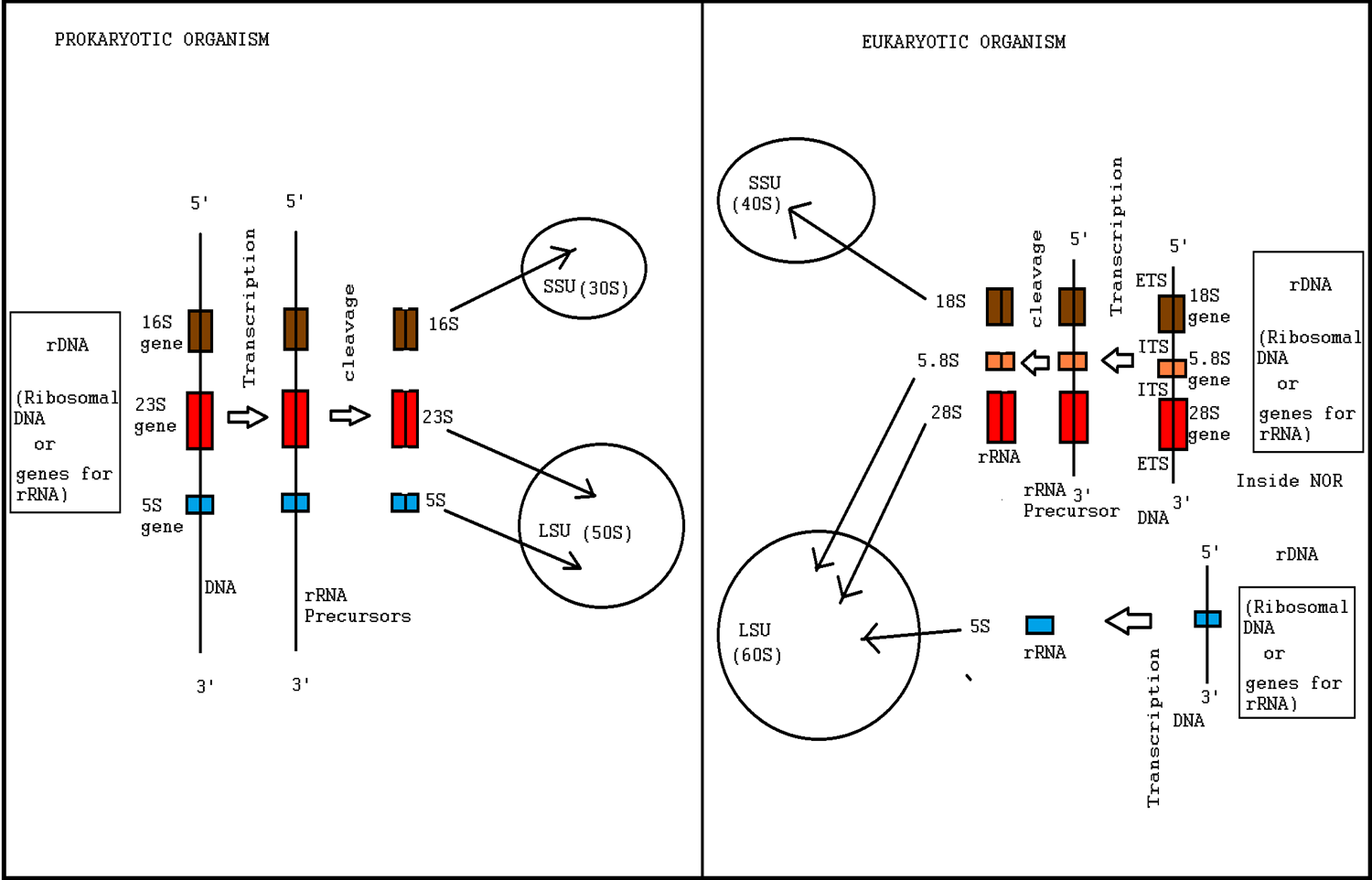
Ribosome Biogenesis
Ribosome biogenesis is the process of making ribosomes. In prokaryotes, this process takes place in the cytoplasm with the transcription of many ribosome gene operons. In eukaryotes, it takes place both in the cytoplasm and in the nucleolus. It involves the coordinated function of over 200 proteins in the synthesis and processing of the three prokaryotic or four eukaryotic rRNAs, as well as assembly of those rRNAs with the ribosomal proteins. Most of the ribosomal proteins fall into various energy-consuming enzyme families including ATP-dependent RNA helicases, AAA-ATPases, GTPases, and kinases. About 60% of a cell's energy is spent on ribosome production and maintenance. Ribosome biogenesis is a very tightly regulated process, and it is closely linked to other cellular activities like growth and division. Some have speculated that in the origin of life, ribosome biogenesis predates cells, and that genes and cells evolved to enhance the reproductive capacity of ribosomes. R ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ribosomal Protein
A ribosomal protein (r-protein or rProtein) is any of the proteins that, in conjunction with rRNA, make up the ribosomal subunits involved in the cellular process of translation. ''E. coli'', other bacteria and Archaea have a 30S small subunit and a 50S large subunit, whereas humans and yeasts have a 40S small subunit and a 60S large subunit. Equivalent subunits are frequently numbered differently between bacteria, Archaea, yeasts and humans. A large part of the knowledge about these organic molecules has come from the study of '' E. coli'' ribosomes. All ribosomal proteins have been isolated and many specific antibodies have been produced. These, together with electronic microscopy and the use of certain reactives, have allowed for the determination of the topography of the proteins in the ribosome. More recently, a near-complete (near)atomic picture of the ribosomal proteins is emerging from the latest high-resolution cryo-EM data (including ). Conservation Ribosomal prot ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ribosomal Protein Leader
A ribosomal protein leader is a mechanism used in cells to control the cellular concentration of a protein that forms a part of the ribosome, and to make sure that the concentration is neither too high nor too low. Ribosomal protein leaders are RNA sequences that are a part of the 5' UTR of mRNAs encoding a ribosomal protein. When cellular concentrations of the ribosomal protein are high, excess protein will bind to the mRNA leader. This binding event can lower gene expression via a number of mechanisms; for example, in the protein-bound state, the RNA could form an intrinsic transcription termination stem-loop. When cellular concentrations of the ribosomal protein are not high, they are occupied in the ribosome, and are not available in significant quantities to bind the mRNA leader. This leads to increased expression of the gene, which leads to the synthesis of more copies of the ribosomal protein. Many examples of ribosomal protein leaders are known in bacteria, including r ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Gram-positive
In bacteriology, gram-positive bacteria are bacteria that give a positive result in the Gram stain test, which is traditionally used to quickly classify bacteria into two broad categories according to their type of cell wall. Gram-positive bacteria take up the crystal violet stain used in the test, and then appear to be purple-coloured when seen through an optical microscope. This is because the thick peptidoglycan layer in the bacterial cell wall retains the stain after it is washed away from the rest of the sample, in the decolorization stage of the test. Conversely, gram-negative bacteria cannot retain the violet stain after the decolorization step; alcohol used in this stage degrades the outer membrane of gram-negative cells, making the cell wall more porous and incapable of retaining the crystal violet stain. Their peptidoglycan layer is much thinner and sandwiched between an inner cell membrane and a bacterial outer membrane, causing them to take up the counterstain (saf ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Bacteria
Bacteria (; singular: bacterium) are ubiquitous, mostly free-living organisms often consisting of one biological cell. They constitute a large domain of prokaryotic microorganisms. Typically a few micrometres in length, bacteria were among the first life forms to appear on Earth, and are present in most of its habitats. Bacteria inhabit soil, water, acidic hot springs, radioactive waste, and the deep biosphere of Earth's crust. Bacteria are vital in many stages of the nutrient cycle by recycling nutrients such as the fixation of nitrogen from the atmosphere. The nutrient cycle includes the decomposition of dead bodies; bacteria are responsible for the putrefaction stage in this process. In the biological communities surrounding hydrothermal vents and cold seeps, extremophile bacteria provide the nutrients needed to sustain life by converting dissolved compounds, such as hydrogen sulphide and methane, to energy. Bacteria also live in symbiotic and parasitic relationsh ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Bacteroidia
Bacteroidales is an order of bacteria. Notably it includes the genera ''Prevotella'' and ''Bacteroides'' , which are commonly found in the human gut microbiota. Phylogeny The currently accepted taxonomy is based on the List of Prokaryotic names with Standing in Nomenclature and National Center for Biotechnology Information (NCBI). Notes See also * List of bacterial orders * List of bacteria genera This article lists the genera of the bacteria. The currently accepted taxonomy is based on the List of Prokaryotic names with Standing in Nomenclature (LPSN) and National Center for Biotechnology Information (NCBI). However many taxonomic names are ... References Bacteroidia {{bacteroidetes-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ribosomal Protein Leader
A ribosomal protein leader is a mechanism used in cells to control the cellular concentration of a protein that forms a part of the ribosome, and to make sure that the concentration is neither too high nor too low. Ribosomal protein leaders are RNA sequences that are a part of the 5' UTR of mRNAs encoding a ribosomal protein. When cellular concentrations of the ribosomal protein are high, excess protein will bind to the mRNA leader. This binding event can lower gene expression via a number of mechanisms; for example, in the protein-bound state, the RNA could form an intrinsic transcription termination stem-loop. When cellular concentrations of the ribosomal protein are not high, they are occupied in the ribosome, and are not available in significant quantities to bind the mRNA leader. This leads to increased expression of the gene, which leads to the synthesis of more copies of the ribosomal protein. Many examples of ribosomal protein leaders are known in bacteria, including r ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |