|
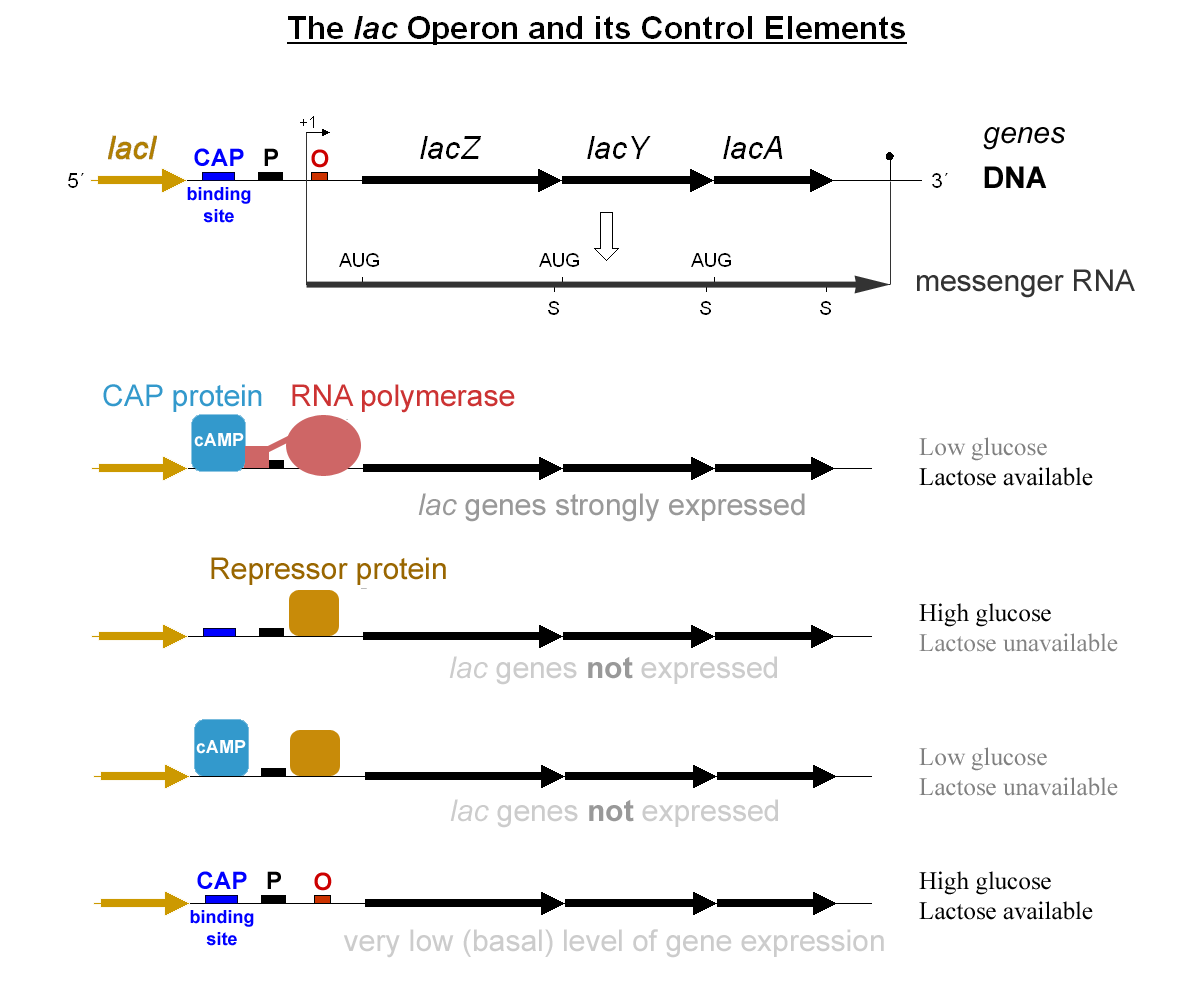
RegulonDB
RegulonDB is a database of the regulatory network of gene expression in ''Escherichia coli'' K-12.{{Cite journal, last=Gama-Castro, first=Socorro, last2=Salgado, first2=Heladia, last3=Santos-Zavaleta, first3=Alberto, last4=Ledezma-Tejeida, first4=Daniela, last5=Muñiz-Rascado, first5=Luis, last6=García-Sotelo, first6=Jair Santiago, last7=Alquicira-Hernández, first7=Kevin, last8=Martínez-Flores, first8=Irma, last9=Pannier, first9=Lucia, date=2016-01-04, title=RegulonDB version 9.0: high-level integration of gene regulation, coexpression, motif clustering and beyond, journal=Nucleic Acids Research, volume=44, issue=D1, pages=D133–143, doi=10.1093/nar/gkv1156, issn=1362-4962, pmc=4702833, pmid=26527724 RegulonDB also models the organization of the genes in transcription units, operons and regulons. A total of 120 sRNAs with 231 total interactions which all together regulate 192 genes are also included. RegulonDB was founded in 1998 and also contributes data to the EcoCyc datab ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Escherichia Coli
''Escherichia coli'' (),Wells, J. C. (2000) Longman Pronunciation Dictionary. Harlow ngland Pearson Education Ltd. also known as ''E. coli'' (), is a Gram-negative, facultative anaerobic, rod-shaped, coliform bacterium of the genus '' Escherichia'' that is commonly found in the lower intestine of warm-blooded organisms. Most ''E. coli'' strains are harmless, but some serotypes ( EPEC, ETEC etc.) can cause serious food poisoning in their hosts, and are occasionally responsible for food contamination incidents that prompt product recalls. Most strains do not cause disease in humans and are part of the normal microbiota of the gut; such strains are harmless or even beneficial to humans (although these strains tend to be less studied than the pathogenic ones). For example, some strains of ''E. coli'' benefit their hosts by producing vitamin K2 or by preventing the colonization of the intestine by pathogenic bacteria. These mutually beneficial relationships between ''E ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Activator (genetics)
A transcriptional activator is a protein (transcription factor) that increases transcription of a gene or set of genes. Activators are considered to have ''positive'' control over gene expression, as they function to promote gene transcription and, in some cases, are required for the transcription of genes to occur. Most activators are DNA-binding proteins that bind to enhancers or promoter-proximal elements. The DNA site bound by the activator is referred to as an "activator-binding site". The part of the activator that makes protein–protein interactions with the general transcription machinery is referred to as an "activating region" or "activation domain". Most activators function by binding sequence-specifically to a regulatory DNA site located near a promoter and making protein–protein interactions with the general transcription machinery ( RNA polymerase and general transcription factors), thereby facilitating the binding of the general transcription machinery to the ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Repressor
In molecular genetics, a repressor is a DNA- or RNA-binding protein that inhibits the expression of one or more genes by binding to the operator or associated silencers. A DNA-binding repressor blocks the attachment of RNA polymerase to the promoter, thus preventing transcription of the genes into messenger RNA. An RNA-binding repressor binds to the mRNA and prevents translation of the mRNA into protein. This blocking or reducing of expression is called repression. Function If an inducer, a molecule that initiates the gene expression, is present, then it can interact with the repressor protein and detach it from the operator. RNA polymerase then can transcribe the message (expressing the gene). A co-repressor is a molecule that can bind to the repressor and make it bind to the operator tightly, which decreases transcription. A repressor that binds with a co-repressor is termed an ''aporepressor'' or ''inactive repressor''. One type of aporepressor is the trp re ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Upstream Activating Sequence
An upstream activating sequence or upstream activation sequence (UAS) is a cis-acting regulatory sequence. It is distinct from the promoter and increases the expression of a neighbouring gene. Due to its essential role in activating transcription, the upstream activating sequence is often considered to be analogous to the function of the enhancer in multicellular eukaryotes. Upstream activation sequences are a crucial part of induction, enhancing the expression of the protein of interest through increased transcriptional activity. The upstream activation sequence is found adjacently upstream to a minimal promoter (TATA box) and serves as a binding site for transactivators. If the transcriptional transactivator does not bind to the UAS in the proper orientation then transcription cannot begin. To further understand the function of an upstream activation sequence, it is beneficial to see its role in the cascade of events that lead to transcription activation. The pathway begins when ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Enhancer (genetics)
In genetics, an enhancer is a short (50–1500 bp) region of DNA that can be bound by proteins ( activators) to increase the likelihood that transcription of a particular gene will occur. These proteins are usually referred to as transcription factors. Enhancers are ''cis''-acting. They can be located up to 1 Mbp (1,000,000 bp) away from the gene, upstream or downstream from the start site. There are hundreds of thousands of enhancers in the human genome. They are found in both prokaryotes and eukaryotes. The first discovery of a eukaryotic enhancer was in the immunoglobulin heavy chain gene in 1983. This enhancer, located in the large intron, provided an explanation for the transcriptional activation of rearranged Vh gene promoters while unrearranged Vh promoters remained inactive. Locations In eukaryotic cells the structure of the chromatin complex of DNA is folded in a way that functionally mimics the supercoiled state characteristic of prokaryotic DNA, so although the e ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
RNA Polymerase
In molecular biology, RNA polymerase (abbreviated RNAP or RNApol), or more specifically DNA-directed/dependent RNA polymerase (DdRP), is an enzyme that synthesizes RNA from a DNA template. Using the enzyme helicase, RNAP locally opens the double-stranded DNA so that one strand of the exposed nucleotides can be used as a template for the synthesis of RNA, a process called transcription. A transcription factor and its associated transcription mediator complex must be attached to a DNA binding site called a promoter region before RNAP can initiate the DNA unwinding at that position. RNAP not only initiates RNA transcription, it also guides the nucleotides into position, facilitates attachment and elongation, has intrinsic proofreading and replacement capabilities, and termination recognition capability. In eukaryotes, RNAP can build chains as long as 2.4 million nucleotides. RNAP produces RNA that, functionally, is either for protein coding, i.e. messenger RNA (mRNA); o ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Terminator (genetics)
In genetics, a transcription terminator is a section of nucleic acid sequence that marks the end of a gene or operon in genomic DNA during transcription. This sequence mediates transcriptional termination by providing signals in the newly synthesized transcript RNA that trigger processes which release the transcript RNA from the transcriptional complex. These processes include the direct interaction of the mRNA secondary structure with the complex and/or the indirect activities of recruited termination factors. Release of the transcriptional complex frees RNA polymerase and related transcriptional machinery to begin transcription of new mRNAs. In prokaryotes Two classes of transcription terminators, Rho-dependent and Rho-independent, have been identified throughout prokaryotic genomes. These widely distributed sequences are responsible for triggering the end of transcription upon normal completion of gene or operon transcription, mediating early termination of transcripts a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Position Weight Matrix
A position weight matrix (PWM), also known as a position-specific weight matrix (PSWM) or position-specific scoring matrix (PSSM), is a commonly used representation of motifs (patterns) in biological sequences. PWMs are often derived from a set of aligned sequences that are thought to be functionally related and have become an important part of many software tools for computational motif discovery. Background Creation Conversion of sequence to position probability matrix A PWM has one row for each symbol of the alphabet (4 rows for nucleotides in DNA sequences or 20 rows for amino acids in protein sequences) and one column for each position in the pattern. In the first step in constructing a PWM, a basic position frequency matrix (PFM) is created by counting the occurrences of each nucleotide at each position. From the PFM, a position probability matrix (PPM) can now be created by dividing that former nucleotide count at each position by the number of sequences, thereb ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Two-component Regulatory System
In the field of molecular biology, a two-component regulatory system serves as a basic stimulus-response coupling mechanism to allow organisms to sense and respond to changes in many different environmental conditions. Two-component systems typically consist of a membrane-bound histidine kinase that senses a specific environmental stimulus and a corresponding response regulator that mediates the cellular response, mostly through differential expression of target genes. Although two-component signaling systems are found in all domains of life, they are most common by far in bacteria, particularly in Gram-negative and cyanobacteria; both histidine kinases and response regulators are among the largest gene families in bacteria. They are much less common in archaea and eukaryotes; although they do appear in yeasts, filamentous fungi, and slime molds, and are common in plants, two-component systems have been described as "conspicuously absent" from animals. Mechanism Two-compon ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Promoter (genetics)
In genetics, a promoter is a sequence of DNA to which proteins bind to initiate transcription of a single RNA transcript from the DNA downstream of the promoter. The RNA transcript may encode a protein (mRNA), or can have a function in and of itself, such as tRNA or rRNA. Promoters are located near the transcription start sites of genes, upstream on the DNA (towards the 5' region of the sense strand). Promoters can be about 100–1000 base pairs long, the sequence of which is highly dependent on the gene and product of transcription, type or class of RNA polymerase recruited to the site, and species of organism. Promoters control gene expression in bacteria and eukaryotes. RNA polymerase must attach to DNA near a gene for transcription to occur. Promoter DNA sequences provide an enzyme binding site. The -10 sequence is TATAAT. -35 sequences are conserved on average, but not in most promoters. Artificial promoters with conserved -10 and -35 elements transcribe more slowly. Al ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Transcription Factor
In molecular biology, a transcription factor (TF) (or sequence-specific DNA-binding factor) is a protein that controls the rate of transcription of genetic information from DNA to messenger RNA, by binding to a specific DNA sequence. The function of TFs is to regulate—turn on and off—genes in order to make sure that they are expressed in the desired cells at the right time and in the right amount throughout the life of the cell and the organism. Groups of TFs function in a coordinated fashion to direct cell division, cell growth, and cell death throughout life; cell migration and organization ( body plan) during embryonic development; and intermittently in response to signals from outside the cell, such as a hormone. There are up to 1600 TFs in the human genome. Transcription factors are members of the proteome as well as regulome. TFs work alone or with other proteins in a complex, by promoting (as an activator), or blocking (as a repressor) the recruitment of ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |