|
RprA RNA
The RprA RNA gene encodes a 106 nucleotide regulatory non-coding RNA. Translational regulation of the stationary phase sigma factor RpoS is mediated by the formation of a double-stranded RNA stem-loop structure in the upstream region of the rpoS messenger RNA, occluding the translation initiation site. Clones carrying rprA (RpoS regulator RNA A) increased the translation of RpoS. As with DsrA, RprA is predicted to form three stem-loops. Thus, at least two small RNAs, DsrA and RprA, participate in the positive regulation of RpoS translation. RprA also appears to bind to the RpoS leader. RprA is non-essential. Wasserman ''et al.'' demonstrated that this RNA is bound by the Hfq protein. Binding to Hfq alters the conformation of RprA. In the presence of Hfq the stability of RprA is influenced by the osmolarity of the cell, this is dependent on the endoribonuclease RNase E. It has been shown the RprA regulates the protein coding genes, called csgD, this protein encodes a stationary ph ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Secondary Structure
Protein secondary structure is the three dimensional form of ''local segments'' of proteins. The two most common secondary structural elements are alpha helices and beta sheets, though beta turns and omega loops occur as well. Secondary structure elements typically spontaneously form as an intermediate before the protein folds into its three dimensional tertiary structure. Secondary structure is formally defined by the pattern of hydrogen bonds between the amino hydrogen and carboxyl oxygen atoms in the peptide backbone. Secondary structure may alternatively be defined based on the regular pattern of backbone dihedral angles in a particular region of the Ramachandran plot regardless of whether it has the correct hydrogen bonds. The concept of secondary structure was first introduced by Kaj Ulrik Linderstrøm-Lang at Stanford in 1952. Other types of biopolymers such as nucleic acids also possess characteristic secondary structures. Types The most common secondary st ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Hfq Protein
The Hfq protein (also known as HF-I protein) encoded by the ''hfq'' gene was discovered in 1968 as an ''Escherichia coli'' host factor that was essential for replication of the bacteriophage Qβ. It is now clear that Hfq is an abundant bacterial RNA binding protein which has many important physiological roles that are usually mediated by interacting with Hfq binding sRNA. In ''E. coli'', Hfq mutants show multiple stress response related phenotypes. The Hfq protein is now known to regulate the translation of two major stress transcription factors ( σS (RpoS) and σE (RpoE) ) in Enterobacteria. It also regulates sRNA in ''Vibrio cholerae'', a specific example being MicX sRNA. In ''Salmonella typhimurium'', Hfq has been shown to be an essential virulence factor as its deletion attenuates the ability of ''S.typhimurium'' to invade epithelial cells, secrete virulence factors or survive in cultured macrophages. In ''Salmonella'', Hfq deletion mutants are also non motile and exhibit ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
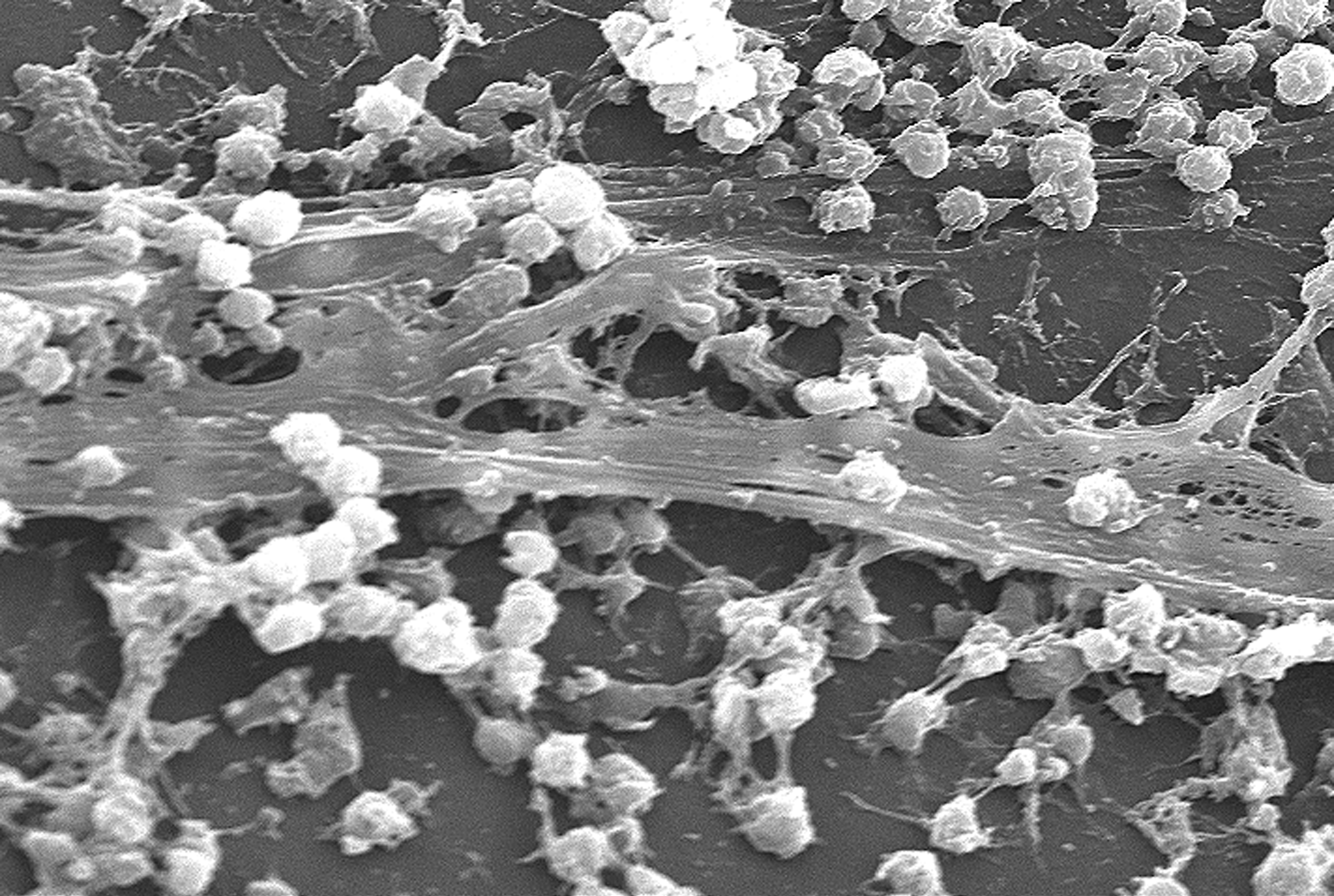
Biofilm
A biofilm comprises any syntrophic consortium of microorganisms in which cells stick to each other and often also to a surface. These adherent cells become embedded within a slimy extracellular matrix that is composed of extracellular polymeric substances (EPSs). The cells within the biofilm produce the EPS components, which are typically a polymeric conglomeration of extracellular polysaccharides, proteins, lipids and DNA. Because they have three-dimensional structure and represent a community lifestyle for microorganisms, they have been metaphorically described as "cities for microbes". Biofilms may form on living or non-living surfaces and can be prevalent in natural, industrial, and hospital settings. They may constitute a microbiome or be a portion of it. The microbial cells growing in a biofilm are physiologically distinct from planktonic cells of the same organism, which, by contrast, are single cells that may float or swim in a liquid medium. Biofilms can form ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
CsgD
CsgD is a transcription and response regulator protein referenced to as the master modulator of bacterial biofilm development. In ''E. coli'' cells, CsgD is tasked with aiding the transition from planktonic cell motility to the stationary phase of biofilm formation, in response to environmental growth factors. A transcription analysis assay (happening ''in vitro'') illustrated a heightened decrease in CsgD's DNA-binding capacity when phosphorylated at A.A. D59 of the protein's primary sequence. Therefore, in the protein's active form (unphosphorylated), CsgD is capable of carrying out its normal functions of regulating curli proteins ( fimbria) and producing ECM polysaccharides (cellulose Cellulose is an organic compound with the formula , a polysaccharide consisting of a linear chain of several hundred to many thousands of β(1→4) linked D-glucose units. Cellulose is an important structural component of the primary cell wall ...). Following a promoter-''lacZ'' fusion ass ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
RNase E
Ribonuclease E is a bacterial ribonuclease that participates in the processing of ribosomal RNA (9S to 5S rRNA) and the chemical degradation of bulk cellular RNA. Cellular localization RNase E was suggested to be a part of the cell membrane protein complex, as it sediments with ribosomes and crude membranes. Microscopy has localized tagged RNase E to the inner cytoplasmic membrane or a helical cytoskeletal structure closely associated with the inner layer. Protein structure This enzyme contains 1,061 residues and separates into two distinct functional regions, which are a large domain located at the 5’N-terminus and a small domain located at the 3’ C-terminus. While N-terminal half forms a catalytic domain, C-terminal half forms a degradosome scaffolding domain. A metal-binding pocket separates them in the middle of the RNase E protein structure. Although degradosome formation does not play a key role for E. coli growth, the deletion of the C-terminal half has bee ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Endoribonuclease
An endoribonuclease is a ribonuclease endonuclease. It cleaves either single-stranded or double-stranded RNA, depending on the enzyme. Example includes both single proteins such as RNase III, RNase A, RNase T1, RNase T2 and RNase H and also complexes of proteins with RNA such as RNase P and the RNA-induced silencing complex. Further examples include endoribonuclease XendoU found in frogs (''Xenopus ''Xenopus'' () (Gk., ξενος, ''xenos''=strange, πους, ''pous''=foot, commonly known as the clawed frog) is a genus of highly aquatic frogs native to sub-Saharan Africa. Twenty species are currently described within it. The two best-kno ...''). External links * {{Nucleases EC 3.1 Ribonucleases ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Osmolarity
Osmotic concentration, formerly known as osmolarity, is the measure of solute concentration, defined as the number of osmoles (Osm) of solute per litre (L) of solution (osmol/L or Osm/L). The osmolarity of a solution is usually expressed as Osm/L (pronounced "osmolar"), in the same way that the molarity of a solution is expressed as "M" (pronounced "molar"). Whereas molarity measures the number of moles of solute per unit volume of solution, osmolarity measures the number of ''osmoles of solute particles'' per unit volume of solution. This value allows the measurement of the osmotic pressure of a solution and the determination of how the solvent will diffuse across a semipermeable membrane (osmosis) separating two solutions of different osmotic concentration. Unit The unit of osmotic concentration is the osmole. This is a non- SI unit of measurement that defines the number of moles of solute that contribute to the osmotic pressure of a solution. A milliosmole (mOsm) is 1 ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DsrA RNA
DsrA RNA is a non-coding RNA that regulates both transcription, by overcoming transcriptional silencing by the nucleoid-associated H-NS protein, and translation, by promoting efficient translation of the stress sigma factor, RpoS. These two activities of DsrA can be separated by mutation: the first of three stem-loops of the 85 nucleotide RNA is necessary for RpoS translation but not for anti-H-NS action, while the second stem-loop is essential for antisilencing and less critical for RpoS translation. The third stem-loop, which behaves as a transcription terminator, can be substituted by the trp transcription terminator without loss of either DsrA function. The sequence of the first stem-loop of DsrA is complementary with the upstream leader portion of RpoS messenger RNA, suggesting that pairing of DsrA with the RpoS message might be important for translational regulation. The structures of DsrA and DsrA/rpoS complex were studied by NMR. The study concluded that the sRNA conta ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Sequence Conservation
In evolutionary biology, conserved sequences are identical or similar Sequence (biology), sequences in nucleic acids (DNA sequence, DNA and RNA) or peptide sequence, proteins across species (homology (biology)#Orthology, orthologous sequences), or within a genome (homology (biology)#Paralogy, paralogous sequences), or between donor and receptor taxa (Sequence homology#Xenology, xenologous sequences). Conservation indicates that a sequence has been maintained by natural selection. A highly conserved sequence is one that has remained relatively unchanged far back up the phylogenetic tree, and hence far back in geological time. Examples of highly conserved sequences include the Ribosomal RNA, RNA components of ribosomes present in all domain (biology), domains of life, the homeobox sequences widespread amongst Eukaryotes, and the tmRNA in Bacteria. The study of sequence conservation overlaps with the fields of genomics, proteomics, evolutionary biology, phylogenetics, bioinformatics ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
RpoS
The gene ''rpoS'' (RNA polymerase, sigma S, also called katF) encodes the sigma factor ''sigma-38'' (σ38, or RpoS), a 37.8 kD protein in ''Escherichia coli''. Sigma factors are proteins that regulate transcription in bacteria. Sigma factors can be activated in response to different environmental conditions. ''rpoS'' is transcribed in late exponential phase, and RpoS is the primary regulator of stationary phase genes. RpoS is a central regulator of the general stress response and operates in both a retroactive and a proactive manner: it not only allows the cell to survive environmental challenges, but it also prepares the cell for subsequent stresses (cross-protection). The transcriptional regulator CsgD is central to biofilm formation, controlling the expression of the curli structural and export proteins, and the diguanylate cyclase, adrA, which indirectly activates cellulose production. The ''rpoS'' gene most likely originated in the gammaproteobacteria. Environmental sig ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Stem-loop
Stem-loop intramolecular base pairing is a pattern that can occur in single-stranded RNA. The structure is also known as a hairpin or hairpin loop. It occurs when two regions of the same strand, usually complementary in nucleotide sequence when read in opposite directions, base-pair to form a double helix that ends in an unpaired loop. The resulting structure is a key building block of many RNA secondary structures. As an important secondary structure of RNA, it can direct RNA folding, protect structural stability for messenger RNA (mRNA), provide recognition sites for RNA binding proteins, and serve as a substrate for enzymatic reactions. Formation and stability The formation of a stem-loop structure is dependent on the stability of the resulting helix and loop regions. The first prerequisite is the presence of a sequence that can fold back on itself to form a paired double helix. The stability of this helix is determined by its length, the number of mismatches or bulges i ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |