|
Reverse Northern Blot
The reverse northern blot is a method by which gene expression patterns may be analyzed by comparing isolated RNA molecules from a tester sample to samples in a control cDNA library. It is a variant of the northern blot in which the nucleic acid immobilized on a membrane is a collection of isolated DNA fragments rather than RNA, and the probe is RNA extracted from a tissue and radioactively labelled. A reverse northern blot can be used to profile expression levels of particular sets of RNA sequences in a tissue or to determine presence of a particular RNA sequence in a sample. Although DNA Microarrays and newer next-generation techniques have generally supplanted reverse northern blotting, it is still utilized today and provides a relatively cheap and easy means of defining expression of large sets of genes. Procedure In order to prepare the reverse northern membrane, cDNA sequences for transcripts of interest are immobilized on nylon membranes, which can be accomplished by use o ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Northern Blot
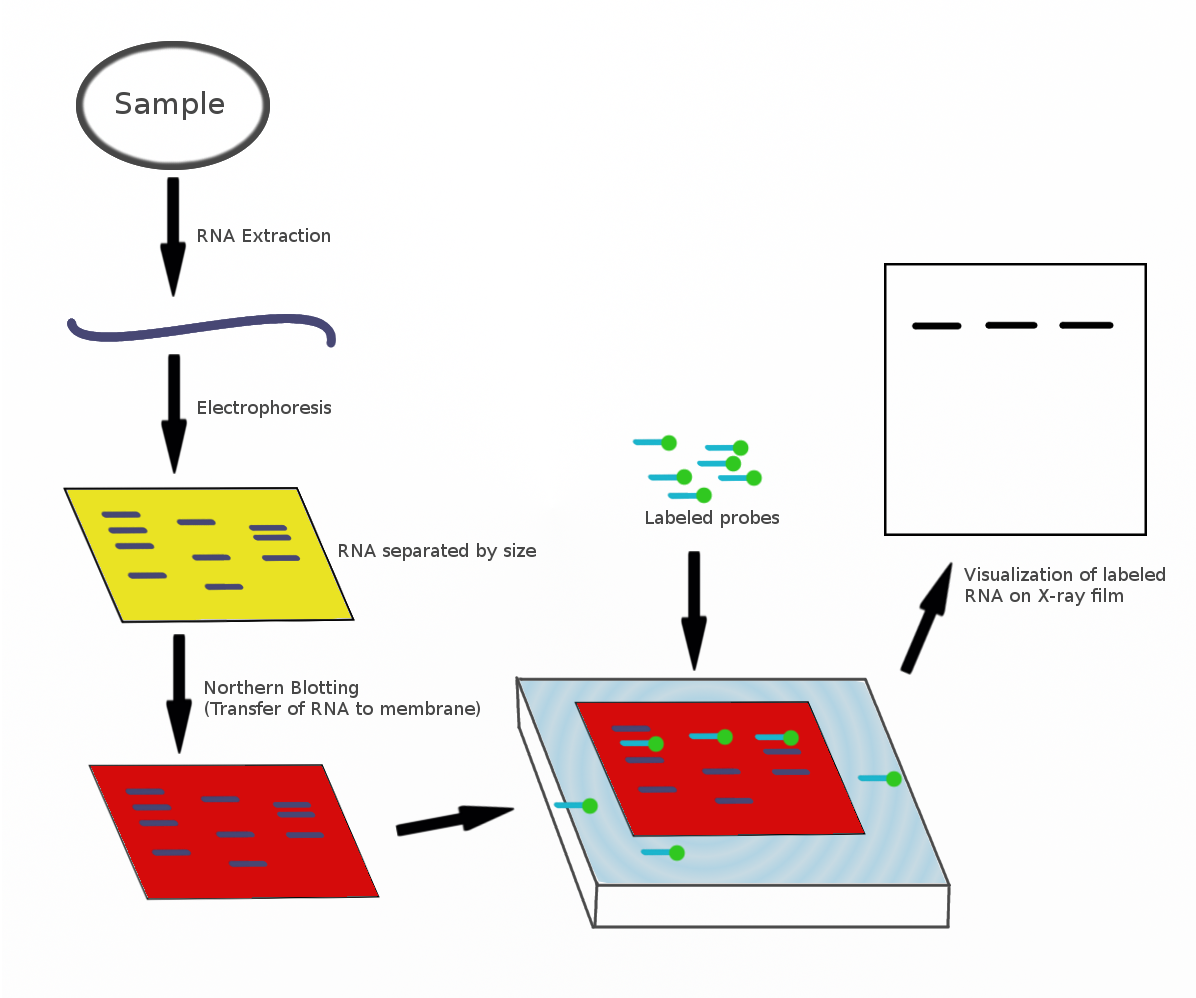
The northern blot, or RNA blot,Gilbert, S. F. (2000) Developmental Biology, 6th Ed. Sunderland MA, Sinauer Associates. is a technique used in molecular biology research to study gene expression by detection of RNA (or isolated mRNA) in a sample.Kevil, C. G., Walsh, L., Laroux, F. S., Kalogeris, T., Grisham, M. B., Alexander, J. S. (1997) An Improved, Rapid Northern Protocol. Biochem. and Biophys. Research Comm. 238:277–279. With northern blotting it is possible to observe cellular control over structure and function by determining the particular gene expression rates during differentiation and morphogenesis, as well as in abnormal or diseased conditions. Northern blotting involves the use of electrophoresis to separate RNA samples by size, and detection with a hybridization probe complementary to part of or the entire target sequence. Strictly speaking, the term 'northern blot' refers specifically to the capillary transfer of RNA from the electrophoresis gel to the blotting m ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Molecular Probe
A molecular probe is a group of atoms or molecules used in molecular biology or chemistry to study the properties of other molecules or structures. If some measurable property of the molecular probe used changes when it interacts with the analyte (such as a change in absorbance), the interactions between the probe and the analyte can be studied. This makes it possible to indirectly study the properties of compounds and structures which may be hard to study directly. The choice of molecular probe will depend on which compound or structure is being studied as well as on what property is of interest. Radioactive DNA or RNA sequences are used in molecular genetics to detect the presence of a complementary sequence by molecular hybridization. Common probes * digoxigenin * ANS * porphyrin * BODIPY * Cyanine * Hybridization probe In molecular biology, a hybridization probe (HP) is a fragment of DNA or RNA of usually 15–10000 nucleotide long which can be radioactively or fluoresc ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Suppression Subtractive Hybridization
Subtractive hybridization is a technology that allows for PCR-based amplification of only cDNA fragments that differ between a control (driver) and experimental transcriptome. cDNA is produced from mRNA. Differences in relative abundance of transcripts are highlighted, as are genetic differences between species. The technique relies on the removal of dsDNA formed by hybridization between a control and test sample, thus eliminating cDNAs or gDNAs of similar abundance, and retaining differentially expressed, or variable in sequence, transcripts or genomic sequences. Suppression subtractive hybridization has also been successfully used to identify strain- or species-specific DNA sequences in a variety of bacteria including ''Vibrio'' species (Metagenomics). See also * Representational difference analysis Representational difference analysis (RDA) is a technique used in biological research to find sequence differences in two genomic or cDNA samples. Genomes or cDNA sequences from ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Differential Display
Differential display (also referred to as DDRT-PCR or DD-PCR) is a laboratory technique that allows a researcher to compare and identify changes in gene expression at the mRNA level between two or more eukaryotic cell samples. It was the most commonly used method to compare expression profiles of two eukaryotic cell samples in the 1990s. By 2000, differential display was superseded by DNA microarray approaches. In differential display, first all the RNA in each sample is reverse transcribed using a set of 3′ "anchored primers" (having a short sequence of deoxy-thymidine nucleotides at the end) to create a cDNA library for each sample, followed by PCR amplification using arbitrary 3′ primers for cDNA strand amplification together with anchored 3′ primers for RNA strand amplification, identical to those used to create the library; about forty arbitrary primers is the optimal number to transcribe almost all of the mRNA. The resulting transcripts are then separated by electrop ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Real-time Polymerase Chain Reaction
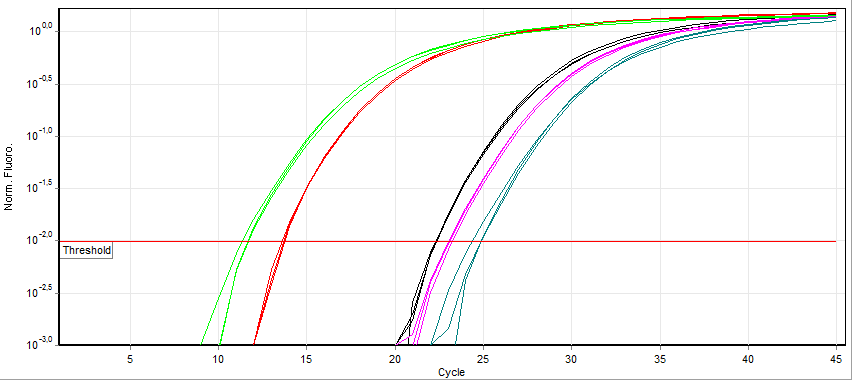
A real-time polymerase chain reaction (real-time PCR, or qPCR) is a laboratory technique of molecular biology based on the polymerase chain reaction (PCR). It monitors the amplification of a targeted DNA molecule during the PCR (i.e., in real time), not at its end, as in conventional PCR. Real-time PCR can be used quantitatively (quantitative real-time PCR) and semi-quantitatively (i.e., above/below a certain amount of DNA molecules) (semi-quantitative real-time PCR). Two common methods for the detection of PCR products in real-time PCR are (1) non-specific fluorescent dyes that intercalate with any double-stranded DNA and (2) sequence-specific DNA probes consisting of oligonucleotides that are labelled with a fluorescent reporter, which permits detection only after hybridization of the probe with its complementary sequence. The Minimum Information for Publication of Quantitative Real-Time PCR Experiments ( MIQE) guidelines propose that the abbreviation ''qPCR'' be used for ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DNA Microarrays
A DNA microarray (also commonly known as DNA chip or biochip) is a collection of microscopic DNA spots attached to a solid surface. Scientists use DNA microarrays to measure the expression levels of large numbers of genes simultaneously or to genotype multiple regions of a genome. Each DNA spot contains picomoles (10−12 moles) of a specific DNA sequence, known as '' probes'' (or ''reporters'' or '' oligos''). These can be a short section of a gene or other DNA element that are used to hybridize a cDNA or cRNA (also called anti-sense RNA) sample (called ''target'') under high-stringency conditions. Probe-target hybridization is usually detected and quantified by detection of fluorophore-, silver-, or chemiluminescence-labeled targets to determine relative abundance of nucleic acid sequences in the target. The original nucleic acid arrays were macro arrays approximately 9 cm × 12 cm and the first computerized image based analysis was published in 1981. It was inv ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Gene Expression Profiling
In the field of molecular biology, gene expression profiling is the measurement of the activity (the expression) of thousands of genes at once, to create a global picture of cellular function. These profiles can, for example, distinguish between cells that are actively dividing, or show how the cells react to a particular treatment. Many experiments of this sort measure an entire genome simultaneously, that is, every gene present in a particular cell. Several transcriptomics technologies can be used to generate the necessary data to analyse. DNA microarrays measure the relative activity of previously identified target genes. Sequence based techniques, like RNA-Seq, provide information on the sequences of genes in addition to their expression level. Background Expression profiling is a logical next step after sequencing a genome: the sequence tells us what the cell could possibly do, while the expression profile tells us what it is actually doing at a point in time. Genes conta ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Gene Expression
Gene expression is the process by which information from a gene is used in the synthesis of a functional gene product that enables it to produce end products, protein or non-coding RNA, and ultimately affect a phenotype, as the final effect. These products are often proteins, but in non-protein-coding genes such as transfer RNA (tRNA) and small nuclear RNA (snRNA), the product is a functional non-coding RNA. Gene expression is summarized in the central dogma of molecular biology first formulated by Francis Crick in 1958, further developed in his 1970 article, and expanded by the subsequent discoveries of reverse transcription and RNA replication. The process of gene expression is used by all known life—eukaryotes (including multicellular organisms), prokaryotes (bacteria and archaea), and utilized by viruses—to generate the macromolecular machinery for life. In genetics, gene expression is the most fundamental level at which the genotype gives rise to the phenotype, '' ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Northern Blot
The northern blot, or RNA blot,Gilbert, S. F. (2000) Developmental Biology, 6th Ed. Sunderland MA, Sinauer Associates. is a technique used in molecular biology research to study gene expression by detection of RNA (or isolated mRNA) in a sample.Kevil, C. G., Walsh, L., Laroux, F. S., Kalogeris, T., Grisham, M. B., Alexander, J. S. (1997) An Improved, Rapid Northern Protocol. Biochem. and Biophys. Research Comm. 238:277–279. With northern blotting it is possible to observe cellular control over structure and function by determining the particular gene expression rates during differentiation and morphogenesis, as well as in abnormal or diseased conditions. Northern blotting involves the use of electrophoresis to separate RNA samples by size, and detection with a hybridization probe complementary to part of or the entire target sequence. Strictly speaking, the term 'northern blot' refers specifically to the capillary transfer of RNA from the electrophoresis gel to the blotting m ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
RNA-Seq
RNA-Seq (named as an abbreviation of RNA sequencing) is a sequencing technique which uses next-generation sequencing (NGS) to reveal the presence and quantity of RNA in a biological sample at a given moment, analyzing the continuously changing cellular transcriptome. Specifically, RNA-Seq facilitates the ability to look at alternative gene spliced transcripts, post-transcriptional modifications, gene fusion, mutations/SNPs and changes in gene expression over time, or differences in gene expression in different groups or treatments. In addition to mRNA transcripts, RNA-Seq can look at different populations of RNA to include total RNA, small RNA, such as miRNA, tRNA, and ribosomal profiling. RNA-Seq can also be used to determine exon/intron boundaries and verify or amend previously annotated 5' and 3' gene boundaries. Recent advances in RNA-Seq include single cell sequencing, in situ sequencing of fixed tissue, and native RNA molecule sequencing with single-molecule real-time ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Southern Blot
A Southern blot is a method used in molecular biology for detection of a specific DNA sequence in DNA samples. Southern blotting combines transfer of electrophoresis-separated DNA fragments to a filter membrane and subsequent fragment detection by probe hybridization. The method is named after the British biologist Edwin Southern, who first published it in 1975. Other blotting methods (i.e., western blot, northern blot, eastern blot, southwestern blot) that employ similar principles, but using RNA or protein, have later been named in reference to Edwin Southern's name. As the label is eponymous, Southern is capitalised, as is conventional of proper nouns. The names for other blotting methods may follow this convention, by analogy. Method #Restriction endonucleases are used to cut high-molecular-weight DNA strands into smaller fragments. #The DNA fragments are then electrophoresed on an agarose gel to separate them by size. # If some of the DNA fragments are larger than ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |



.jpg)