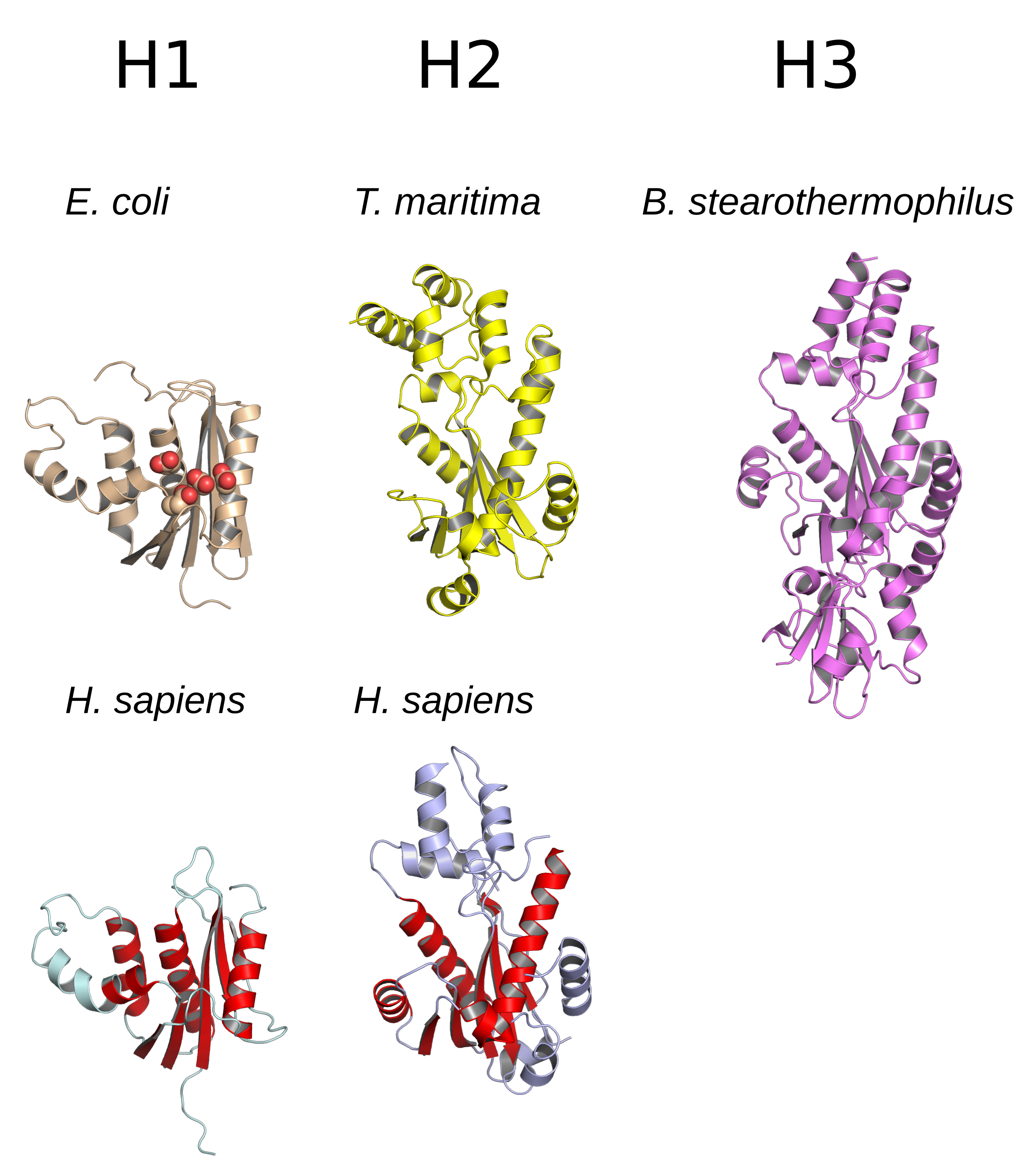
|
Retropepsin
Retroviral aspartyl proteases or retropepsins are single domain aspartyl proteases from retroviruses, retrotransposons, and badnaviruses (plant dsDNA viruses). These proteases are generally part of a larger pol or gag polyprotein. Retroviral proteases are homologous to a single domain of the two-domain eukaryotic aspartyl proteases such as pepsins, cathepsins, and renins (; MEROPS A1). Retropepsins are members of MEROPS A2, clan AA. All known members are endopeptidases. The enzyme is only active as a homodimer, as each one corresponds to half of the eukaryotic two-lobe enzyme. The two parts each contribute one catalytic aspartyl residue. Retroviral aspartyl protease is synthesised as part of the pol polyprotein that contains an aspartyl protease, a reverse transcriptase, RNase H and integrase. pol polyprotein undergoes specific enzymatic cleavage to yield the mature proteins. Not all retroviral aspartyl proteases generated from ''pol'' are retropepsins in the stric ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
HIV-1 Protease
HIV-1 protease (PR) is a retroviral aspartyl protease (retropepsin), an enzyme involved with peptide bond hydrolysis in retroviruses, that is essential for the life-cycle of HIV, the retrovirus that causes AIDS. HIV protease cleaves newly synthesized polyproteins (namely, Gag and Gag- Pol) at nine cleavage sites to create the mature protein components of an HIV virion, the infectious form of a virus outside of the host cell. Without effective HIV protease, HIV virions remain uninfectious. Structure Mature HIV protease exists as a 22 kDa homodimer, with each subunit made up of 99 amino acids. A single active site lies between the identical subunits and has the characteristic Asp- Thr- Gly (Asp25, Thr26 and Gly27) catalytic triad sequence common to aspartic proteases. As HIV-1 PR can only function as a dimer, the mature protease contains two Asp25 amino acids, one from each monomer, that act in conjunction with each other as the catalytic residues. Additionally, HIV protea ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Dimer
In biochemistry, a protein dimer is a macromolecular complex formed by two protein monomers, or single proteins, which are usually non-covalently bound. Many macromolecules, such as proteins or nucleic acids, form dimers. The word ''dimer'' has roots meaning "two parts", '' di-'' + '' -mer''. A protein dimer is a type of protein quaternary structure. A protein homodimer is formed by two identical proteins. A protein heterodimer is formed by two different proteins. Most protein dimers in biochemistry are not connected by covalent bonds. An example of a non-covalent heterodimer is the enzyme reverse transcriptase, which is composed of two different amino acid chains. An exception is dimers that are linked by disulfide bridges such as the homodimeric protein NEMO. Some proteins contain specialized domains to ensure dimerization (dimerization domains) and specificity. The G protein-coupled cannabinoid receptors have the ability to form both homo- and heterodimers with several ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Endopeptidases
Endopeptidase or endoproteinase are proteolytic peptidases that break peptide bonds of nonterminal amino acids (i.e. within the molecule), in contrast to exopeptidases, which break peptide bonds from end-pieces of terminal amino acids. For this reason, endopeptidases cannot break down peptides into monomers, while exopeptidases can break down proteins into monomers. A particular case of endopeptidase is the oligopeptidase, whose substrates are oligopeptides instead of proteins. They are usually very specific for certain amino acids. Examples of endopeptidases include: * Trypsin - cuts after Arg or Lys, unless followed by Pro. Very strict. Works best at pH 8. * Chymotrypsin - cuts after Phe, Trp, or Tyr, unless followed by Pro. Cuts more slowly after His, Met or Leu. Works best at pH 8. * Elastase - cuts after Ala, Gly, Ser, or Val, unless followed by Pro. * Thermolysin - cuts ''before'' Ile, Met, Phe, Trp, Tyr, or Val, unless ''preceded'' by Pro. Sometimes cuts after Ala, Asp, ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Proteases
A protease (also called a peptidase, proteinase, or proteolytic enzyme) is an enzyme that catalyzes (increases reaction rate or "speeds up") proteolysis, breaking down proteins into smaller polypeptides or single amino acids, and spurring the formation of new protein products. They do this by cleaving the peptide bonds within proteins by hydrolysis, a reaction where water breaks bonds. Proteases are involved in many biological functions, including digestion of ingested proteins, protein catabolism (breakdown of old proteins), and cell signaling. In the absence of functional accelerants, proteolysis would be very slow, taking hundreds of years. Proteases can be found in all forms of life and viruses. They have independently evolved multiple times, and different classes of protease can perform the same reaction by completely different catalytic mechanisms. Hierarchy of proteases Based on catalytic residue Proteases can be classified into seven broad groups: * Serine proteases - ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
ERVK6
HERV-K_19q12 provirus ancestral Pol protein is a protein that in humans is encoded by the ''ERVK6'' gene. References Further reading * * * * * * * * * * * * * * * * * Genes Human proteins {{protein-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DDI2
DDI may stand for: Companies and organizations * DD International, international TV channel in India * Development Dimensions International, a talent management company * Direct Democracy Ireland, a political party in Ireland * KDDI, formerly DDI, a Japanese telecommunications company Science * Drug–drug interaction * Didanosine, an antiretroviral drug Technology * Acronym for DNS/DHCP/IPAM; see IP Address Management * Direct dial-in, or direct inward dialing, a telecommunications service * ''D&D'' Insider, an online method used to deliver ''Dungeons & Dragons'' content * Diverging diamond interchange, a road structure that guides traffic Other uses * Dance Dance Immolation, interactive performance piece * Data Documentation Initiative, a standard for describing surveys, questionnaires, and statistical data files * Democracy-Dictatorship Index, a binary measure of democracy and dictatorship * Divisional detective inspector {{unreferenced, date=October 2008 Divisional det ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DDI1
DNA-damage inducible 1 homolog 1 (S. cerevisiae) is a protein. In humans it is encoded by the DDI1 gene In biology, the word gene (from , ; "...Wilhelm Johannsen coined the word gene to describe the Mendelian units of heredity..." meaning ''generation'' or ''birth'' or ''gender'') can have several different meanings. The Mendelian gene is a ba .... References Further reading External links PDBe-KBprovides an overview of all the structure information available in the PDB for Human Protein DDI1 homolog 1 Human proteins {{gene-11-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Spumavirus
''Spumaretrovirinae'', commonly called spumaviruses (, Latin for "foam") or foamyviruses, is a subfamily of the ''Retroviridae'' family. ICTVMaster Species List 2018a v1MSL including all taxa updates since the 2017 release. Fall 2018 (MSL #33) Spumaviruses are exogenous viruses that have specific morphology with prominent surface spikes. The virions contain significant amounts of double-stranded full-length DNA, and assembly is rather unusual in these viruses. Spumaviruses are unlike most enveloped viruses in that the envelope membrane is acquired by budding through the endoplasmic reticulum instead of the cytoplasmic membrane. Some spumaviruses, including the equine foamy virus (EFV), bud from the cytoplasmic membrane. Some examples of these viruses are simian foamy virus and the human foamy virus Human foamy virus (HFV) is a retrovirus and specifically belongs to the genus ''Spumavirus''. The spumaviruses are complex and significantly different from the other six genera ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Integrase
Retroviral integrase (IN) is an enzyme produced by a retrovirus (such as HIV) that integrates—forms covalent links between—its genetic information into that of the host cell it infects. Retroviral INs are not to be confused with phage integrases (recombinases) used in biotechnology, such as λ phage integrase, as discussed in site-specific recombination. The macromolecular complex of an IN macromolecule bound to the ends of the viral DNA ends has been referred to as the '' intasome''; IN is a key component in this and the retroviral pre-integration complex. Structure All retroviral IN proteins contain three canonical domains, connected by flexible linkers: ● an N-terminal HH-CC zinc-binding domain (a three-helical bundle stabilized by coordination of a Zn(II) cation) ● a catalytic core domain (RNaseH fold) ● a C-terminal DNA-binding domain ( SH3 fold). Crystal and NMR structures of the individual domains and 2-domain constructs of integrases ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
RNase H
Ribonuclease H (abbreviated RNase H or RNH) is a family of non-sequence-specific endonuclease enzymes that catalyze the cleavage of RNA in an RNA/ DNA substrate via a hydrolytic mechanism. Members of the RNase H family can be found in nearly all organisms, from bacteria to archaea to eukaryotes. The family is divided into evolutionarily related groups with slightly different substrate preferences, broadly designated ribonuclease H1 and H2. The human genome encodes both H1 and H2. Human ribonuclease H2 is a heterotrimeric complex composed of three subunits, mutations in any of which are among the genetic causes of a rare disease known as Aicardi–Goutières syndrome. A third type, closely related to H2, is found only in a few prokaryotes, whereas H1 and H2 occur in all domains of life. Additionally, RNase H1-like retroviral ribonuclease H domains occur in multidomain reverse transcriptase proteins, which are encoded by retroviruses such as HIV and are required for viral repl ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Reverse Transcriptase
A reverse transcriptase (RT) is an enzyme used to generate complementary DNA (cDNA) from an RNA template, a process termed reverse transcription. Reverse transcriptases are used by viruses such as HIV and hepatitis B to replicate their genomes, by retrotransposon mobile genetic elements to proliferate within the host genome, and by eukaryotic cells to extend the telomeres at the ends of their linear chromosomes. Contrary to a widely held belief, the process does not violate the flows of genetic information as described by the classical central dogma, as transfers of information from RNA to DNA are explicitly held possible. Retroviral RT has three sequential biochemical activities: RNA-dependent DNA polymerase activity, ribonuclease H (RNase H), and DNA-dependent DNA polymerase activity. Collectively, these activities enable the enzyme to convert single-stranded RNA into double-stranded cDNA. In retroviruses and retrotransposons, this cDNA can then integrate into the host genom ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Homodimer
In biochemistry, a protein dimer is a macromolecular complex formed by two protein monomers, or single proteins, which are usually non-covalently bound. Many macromolecules, such as proteins or nucleic acids, form dimers. The word ''dimer'' has roots meaning "two parts", '' di-'' + '' -mer''. A protein dimer is a type of protein quaternary structure. A protein homodimer is formed by two identical proteins. A protein heterodimer is formed by two different proteins. Most protein dimers in biochemistry are not connected by covalent bonds. An example of a non-covalent heterodimer is the enzyme reverse transcriptase, which is composed of two different amino acid chains. An exception is dimers that are linked by disulfide bridges such as the homodimeric protein NEMO. Some proteins contain specialized domains to ensure dimerization (dimerization domains) and specificity. The G protein-coupled cannabinoid receptors have the ability to form both homo- and heterodimers with several ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |