|
RefDB (chemistry)
The Re-referenced Protein Chemical shift Database (RefDB) is an NMR spectroscopy database of carefully corrected or re-referenced chemical shifts, derived from the BioMagResBank (BMRB) (Fig. 1). The database was assembled by using a structure-based chemical shift calculation program (called SHIFTX) to calculate expected protein (1)H, (13)C and (15)N chemical shifts from X-ray or NMR coordinate data of previously assigned proteins reported in the BMRB. The comparison is automatically performed by a program called SHIFTCOR. The RefDB database currently provides reference-corrected chemical shift data on more than 2000 assigned peptides and proteins. Data from the database indicates that nearly 25% of BMRB entries with (13)C protein assignments and 27% of BMRB entries with (15)N protein assignments require significant chemical shift reference readjustments. Additionally, nearly 40% of protein entries deposited in the BioMagResBank appear to have at least one assignment error. Users ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
David S
David (; , "beloved one") (traditional spelling), , ''Dāwūd''; grc-koi, Δαυΐδ, Dauíd; la, Davidus, David; gez , ዳዊት, ''Dawit''; xcl, Դաւիթ, ''Dawitʿ''; cu, Давíдъ, ''Davidŭ''; possibly meaning "beloved one". was, according to the Hebrew Bible, the third king of the United Kingdom of Israel. In the Books of Samuel, he is described as a young shepherd and harpist who gains fame by slaying Goliath, a champion of the Philistines, in southern Canaan. David becomes a favourite of Saul, the first king of Israel; he also forges a notably close friendship with Jonathan, a son of Saul. However, under the paranoia that David is seeking to usurp the throne, Saul attempts to kill David, forcing the latter to go into hiding and effectively operate as a fugitive for several years. After Saul and Jonathan are both killed in battle against the Philistines, a 30-year-old David is anointed king over all of Israel and Judah. Following his rise to power, Da ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Chemical Shift
In nuclear magnetic resonance (NMR) spectroscopy, the chemical shift is the resonant frequency of an atomic nucleus relative to a standard in a magnetic field. Often the position and number of chemical shifts are diagnostic of the structure of a molecule. Chemical shifts are also used to describe signals in other forms of spectroscopy such as photoemission spectroscopy. Some atomic nuclei possess a magnetic moment ( nuclear spin), which gives rise to different energy levels and resonance frequencies in a magnetic field. The total magnetic field experienced by a nucleus includes local magnetic fields induced by currents of electrons in the molecular orbitals (note that electrons have a magnetic moment themselves). The electron distribution of the same type of nucleus (e.g. ) usually varies according to the local geometry (binding partners, bond lengths, angles between bonds, and so on), and with it the local magnetic field at each nucleus. This is reflected in the spin energy le ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Methods
Protein methods are the techniques used to study proteins. There are experimental methods for studying proteins (e.g., for detecting proteins, for isolating and purifying proteins, and for characterizing the structure and function of proteins, often requiring that the protein first be purified). Computational methods typically use computer programs to analyze proteins. However, many experimental methods (e.g., mass spectrometry) require computational analysis of the raw data. Genetic methods Experimental analysis of proteins typically requires expression and purification of proteins. Expression is achieved by manipulating DNA that encodes the protein(s) of interest. Hence, protein analysis usually requires DNA methods, especially cloning. Some examples of genetic methods include conceptual translation, Site-directed mutagenesis, using a fusion protein, and matching allele with disease states. Some proteins have never been directly sequenced, however by translating codons from kn ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Nuclear Magnetic Resonance Software
Nuclear may refer to: Physics Relating to the nucleus of the atom: *Nuclear engineering * Nuclear physics * Nuclear power * Nuclear reactor * Nuclear weapon * Nuclear medicine * Radiation therapy * Nuclear warfare Mathematics * Nuclear space * Nuclear operator * Nuclear congruence * Nuclear C*-algebra Biology Relating to the nucleus of the cell: * Nuclear DNA Society *Nuclear family, a family consisting of a pair of adults and their children Music * "Nuclear" (band), group music. * "Nuclear" (Ryan Adams song), 2002 *"Nuclear", a song by Mike Oldfield from his '' Man on the Rocks'' album * ''Nu.Clear'' (EP) by South Korean girl group CLC See also * Nucleus (other) * Nucleolus *Nucleation *Nucleic acid Nucleic acids are biopolymers, macromolecules, essential to all known forms of life. They are composed of nucleotides, which are the monomers made of three components: a 5-carbon sugar, a phosphate group and a nitrogenous base. The two main cl ... * Nucular ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Chemical Databases
A chemical database is a database specifically designed to store chemical information. This information is about chemical and crystal structures, spectra, reactions and syntheses, and thermophysical data. Types of chemical databases Bioactivity database Bioactivity databases correlate structures or other chemical information to bioactivity results taken from bioassays in literature, patents, and screening programs. Chemical structures Chemical structures are traditionally represented using lines indicating chemical bonds between atoms and drawn on paper (2D structural formulae). While these are ideal visual representations for the chemist, they are unsuitable for computational use and especially for search and storage. Small molecules (also called ligands in drug design applications), are usually represented using lists of atoms and their connections. Large molecules such as proteins are however more compactly represented using the sequences of their amino acid bui ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein NMR
Nuclear magnetic resonance spectroscopy of proteins (usually abbreviated protein NMR) is a field of structural biology in which NMR spectroscopy is used to obtain information about the structure and dynamics of proteins, and also nucleic acids, and their complexes. The field was pioneered by Richard R. Ernst and Kurt Wüthrich at the ETH, and by Ad Bax, Marius Clore, Angela Gronenborn at the National Institutes of Health, NIH, and Gerhard Wagner (physicist), Gerhard Wagner at Harvard University, among others. Structure determination by NMR spectroscopy usually consists of several phases, each using a separate set of highly specialized techniques. The sample is prepared, measurements are made, interpretive approaches are applied, and a structure is calculated and validated. NMR involves the quantum-mechanical properties of the central core ("Atomic nucleus, nucleus") of the atom. These properties depend on the local molecular environment, and their measurement provides a map of how th ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
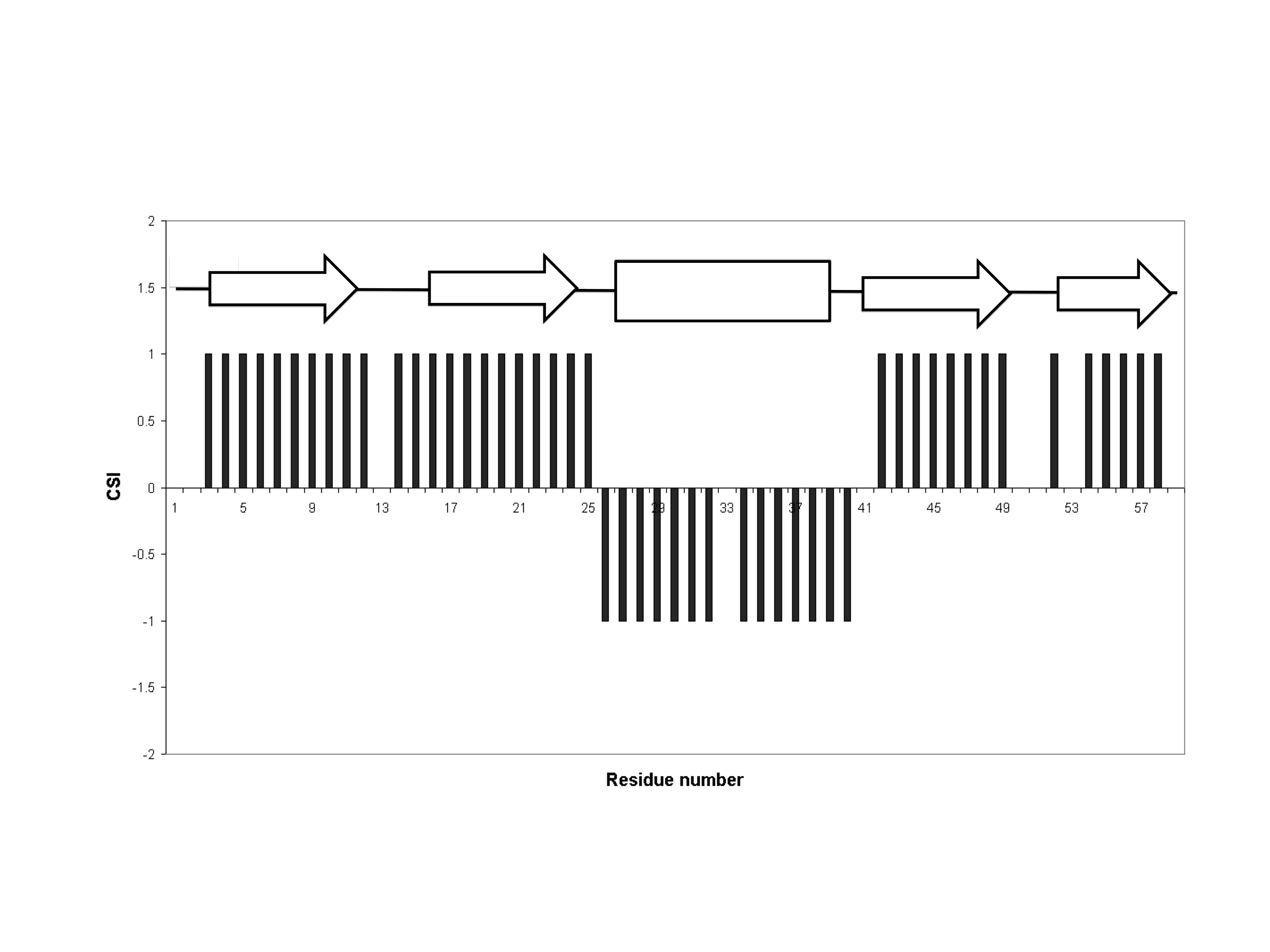
Chemical Shift Index
The chemical shift index or CSI is a widely employed technique in protein nuclear magnetic resonance spectroscopy that can be used to display and identify the location (i.e. start and end) as well as the type of protein secondary structure (beta strands, helices and random coil regions) found in proteins using only backbone chemical shift data The technique was invented by David S. Wishart in 1992 for analyzing 1Hα chemical shifts and then later extended by him in 1994 to incorporate 13C backbone shifts. The original CSI method makes use of the fact that 1Hα chemical shifts of amino acid residues in helices tends to be shifted upfield (i.e. towards the right side of an NMR spectrum) relative to their random coil values and downfield (i.e. towards the left side of an NMR spectrum) in beta strands. Similar kinds of upfield and downfield trends are also detectable in backbone 13C chemical shifts. Implementation The CSI is a graph-based technique that essentially employs an amino ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Secondary Structure
Protein secondary structure is the three dimensional form of ''local segments'' of proteins. The two most common secondary structural elements are alpha helices and beta sheets, though beta turns and omega loops occur as well. Secondary structure elements typically spontaneously form as an intermediate before the protein folds into its three dimensional tertiary structure. Secondary structure is formally defined by the pattern of hydrogen bonds between the amino hydrogen and carboxyl oxygen atoms in the peptide backbone. Secondary structure may alternatively be defined based on the regular pattern of backbone dihedral angles in a particular region of the Ramachandran plot regardless of whether it has the correct hydrogen bonds. The concept of secondary structure was first introduced by Kaj Ulrik Linderstrøm-Lang at Stanford in 1952. Other types of biopolymers such as nucleic acids also possess characteristic secondary structures. Types The most common secondary ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Structure Database
In biology, a protein structure database is a database that is modeled around the various experimentally determined protein structures. The aim of most protein structure databases is to organize and annotate the protein structures, providing the biological community access to the experimental data in a useful way. Data included in protein structure databases often includes three-dimensional coordinates as well as experimental information, such as unit cell dimensions and angles for x-ray crystallography determined structures. Though most instances, in this case either proteins or a specific structure determinations of a protein, also contain sequence information and some databases even provide means for performing sequence based queries, the primary attribute of a structure database is structural information, whereas sequence databases focus on sequence information, and contain no structural information for the majority of entries. Protein structure databases are critical for man ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Backbone Chain
In polymer science, the polymer chain or simply backbone of a polymer is the main chain of a polymer. Polymers are often classified according to the elements in the main chains. The character of the backbone, i.e. its flexibility, determines the properties of the polymer (such as the glass transition temperature). For example, in polysiloxanes (silicone), the backbone chain is very flexible, which results in a very low glass transition temperature of . The polymers with rigid backbones are prone to crystallization (e.g. polythiophenes) in thin films and in solution. Crystallization in its turn affects the optical properties of the polymers, its optical band gap and electronic levels. Organic polymers : Common synthetic polymers have main chains composed of carbon, i.e. C-C-C-C.... Examples include polyolefins such as polyethylene ((CH2CH2)n) and many substituted derivative ((CH2CH(R))n) such as polystyrene (R = C6H5), polypropylene (R = CH3), and acrylates (R = CO2R'). Other ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |