|
QpGraph
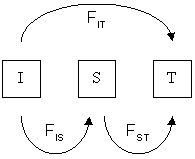
ADMIXTOOLS (or AdmixTools) is an R software package that is primarily used for analyzing admixture in population genetics. The initial version was originally developed by Nick Patterson and colleagues in 2012. The second reimplemented version, ADMIXTOOLS 2, which has a revised point-and-click interface and improved speed, was developed jointly by Nick Patterson and David Reich. qpGraph qpGraph is a software program that is part of the ADMIXTOOLS software package developed by Patterson et al. (2012). qpGraph builds graphs that model genetic admixture in population relationships. It estimates network topology likelihoods from a fixed number of admixture events, based on the observed f-statistics. Other tools Related statistical tools in the ADMIXTOOLS software package include qpAdm, qpDstat, and qpWave. qpDstat and qpWave test to see if populations form clades, while qpAdm estimates ancestry proportions. See also *SplitsTree *Dendroscope *List of phylogenetics software Refere ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Nick Patterson (scientist)
Nicholas James Patterson (born 9 June 1947) is a mathematician working as a staff scientist at the Broad Institute with notable contributions to the area of computational genomics. His work has appeared in scientific journals such as ''Nature'', ''Science'' and ''Nature Genetics''. His research has brought a better understanding of early human migrations. He is among the group of scientists who have sequenced the Neanderthal genome in 2010. This was followed by the sequencing of a much higher quality Neanderthal genome, where the subject was from the Altai Mountains, in 2014. These studies have uncovered some unexpected facts about the interbreeding between archaic and modern humans. Biography Patterson was an only child who grew up in the Bayswater section of central London. Patterson received his B.A. and Ph.D. in mathematics at Cambridge University. His doctoral advisor was John G. Thompson. He initially worked for the British code-breaking agency GCHQ and the Center ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
David Reich (geneticist)
David Emil Reich (born July 14, 1974) is an American geneticist known for his research into the population genetics of ancient humans, including their migrations and the mixing of populations, discovered by analysis of genome-wide patterns of mutations. He is professor in the department of genetics at the Harvard Medical School, and an associate of the Broad Institute. Reich was highlighted as one of Nature's 10 for his contributions to science in 2015. He received the Dan David Prize in 2017, the NAS Award in Molecular Biology, the Wiley Prize, and the Darwin–Wallace Medal in 2019. In 2021 he was awarded the Massry Prize. Early life Reich grew up as part of a Jewish family in Washington, D.C. His parents are novelist Tova Reich (sister of Rabbi Avi Weiss) and Walter Reich, a professor at George Washington University, who served as the first director of the United States Holocaust Memorial Museum. David Reich started out as a sociology major as an undergraduate at Harvard Colle ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
R (programming Language)
R is a programming language for statistical computing and graphics supported by the R Core Team and the R Foundation for Statistical Computing. Created by statisticians Ross Ihaka and Robert Gentleman, R is used among data miners, bioinformaticians and statisticians for data analysis and developing statistical software. Users have created packages to augment the functions of the R language. According to user surveys and studies of scholarly literature databases, R is one of the most commonly used programming languages used in data mining. R ranks 12th in the TIOBE index, a measure of programming language popularity, in which the language peaked in 8th place in August 2020. The official R software environment is an open-source free software environment within the GNU package, available under the GNU General Public License. It is written primarily in C, Fortran, and R itself (partially self-hosting). Precompiled executables are provided for various operating systems. R ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Population Genetics
Population genetics is a subfield of genetics that deals with genetic differences within and between populations, and is a part of evolutionary biology. Studies in this branch of biology examine such phenomena as adaptation, speciation, and population structure. Population genetics was a vital ingredient in the emergence of the modern evolutionary synthesis. Its primary founders were Sewall Wright, J. B. S. Haldane and Ronald Fisher, who also laid the foundations for the related discipline of quantitative genetics. Traditionally a highly mathematical discipline, modern population genetics encompasses theoretical, laboratory, and field work. Population genetic models are used both for statistical inference from DNA sequence data and for proof/disproof of concept. What sets population genetics apart from newer, more phenotypic approaches to modelling evolution, such as evolutionary game theory and adaptive dynamics, is its emphasis on such genetic phenomena as dominance, epi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Population Genetics
Population genetics is a subfield of genetics that deals with genetic differences within and between populations, and is a part of evolutionary biology. Studies in this branch of biology examine such phenomena as adaptation, speciation, and population structure. Population genetics was a vital ingredient in the emergence of the modern evolutionary synthesis. Its primary founders were Sewall Wright, J. B. S. Haldane and Ronald Fisher, who also laid the foundations for the related discipline of quantitative genetics. Traditionally a highly mathematical discipline, modern population genetics encompasses theoretical, laboratory, and field work. Population genetic models are used both for statistical inference from DNA sequence data and for proof/disproof of concept. What sets population genetics apart from newer, more phenotypic approaches to modelling evolution, such as evolutionary game theory and adaptive dynamics, is its emphasis on such genetic phenomena as dominance, epi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Point-and-click
Point and click are the actions of a computer user moving a pointer to a certain location on a screen (''pointing'') and then pressing a button on a mouse, usually the left button (''click''), or other pointing device. An example of point and click is in hypermedia, where users click on hyperlinks to navigate from document to document. Point and click can be used with any number of input devices varying from mouses, touch pads, trackpoint, joysticks, scroll buttons, and roller balls. User interfaces, for example graphical user interfaces, are sometimes described as "point-and-click interfaces", often to suggest that they are very easy to use, requiring that the user simply point to indicate their wishes. These interfaces are sometimes referred to condescendingly (e.g., by Unix users) as "click-and-drool" or "point-and-drool" interfaces. The use of this phrase to describe software implies that the interface can be controlled solely through the mouse (or some other means such as ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Genetic Admixture
Genetic admixture occurs when previously diverged or isolated genetic lineages mix.⅝ Admixture results in the introduction of new genetic lineages into a population. Examples Climatic cycles facilitate genetic admixture in cold periods and genetic diversification in warm periods. Natural flooding can cause genetic admixture within populations of migrating fish species. Genetic admixture may have an important role for the success of populations that colonise a new area and interbreed with individuals of native populations. Mapping Admixture mapping is a method of gene mapping that uses a population of mixed ancestry (an admixed population) to find the genetic loci that contribute to differences in diseases or other phenotypes found between the different ancestral populations. The method is best applied to populations with recent admixture from two populations that were previously genetically isolated. The method attempts to correlate the degree of ancestry near a genetic locus w ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Likelihood Function
The likelihood function (often simply called the likelihood) represents the probability of random variable realizations conditional on particular values of the statistical parameters. Thus, when evaluated on a given sample, the likelihood function indicates which parameter values are more ''likely'' than others, in the sense that they would have made the observed data more probable. Consequently, the likelihood is often written as \mathcal(\theta\mid X) instead of P(X \mid \theta), to emphasize that it is to be understood as a function of the parameters \theta instead of the random variable X. In maximum likelihood estimation, the arg max of the likelihood function serves as a point estimate for \theta, while local curvature (approximated by the likelihood's Hessian matrix) indicates the estimate's precision. Meanwhile in Bayesian statistics, parameter estimates are derived from the converse of the likelihood, the so-called posterior probability, which is calculated via Bayes' r ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
F-statistics
In population genetics, ''F''-statistics (also known as fixation indices) describe the statistically expected level of heterozygosity in a population; more specifically the expected degree of (usually) a reduction in heterozygosity when compared to Hardy–Weinberg expectation. ''F''-statistics can also be thought of as a measure of the correlation between genes drawn at different levels of a (hierarchically) subdivided population. This correlation is influenced by several evolutionary processes, such as genetic drift, founder effect, bottleneck, genetic hitchhiking, meiotic drive, mutation, gene flow, inbreeding, natural selection, or the Wahlund effect, but it was originally designed to measure the amount of allelic fixation owing to genetic drift. The concept of ''F''-statistics was developed during the 1920s by the American geneticist Sewall Wright, who was interested in inbreeding in cattle. However, because complete dominance causes the phenotypes of homozygote dominants ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Clade
A clade (), also known as a monophyletic group or natural group, is a group of organisms that are monophyletic – that is, composed of a common ancestor and all its lineal descendants – on a phylogenetic tree. Rather than the English term, the equivalent Latin term ''cladus'' (plural ''cladi'') is often used in taxonomical literature. The common ancestor may be an individual, a population, or a species (extinct or extant). Clades are nested, one in another, as each branch in turn splits into smaller branches. These splits reflect evolutionary history as populations diverged and evolved independently. Clades are termed monophyletic (Greek: "one clan") groups. Over the last few decades, the cladistic approach has revolutionized biological classification and revealed surprising evolutionary relationships among organisms. Increasingly, taxonomists try to avoid naming taxa that are not clades; that is, taxa that are not monophyletic. Some of the relationships between organisms ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
SplitsTree
SplitsTree is a popular freeware program for inferring phylogenetic trees, phylogenetic networks, or, more generally, splits graphs, from various types of data such as a sequence alignment, a distance matrix or a set of trees. SplitsTree implements published methods such as split decomposition, neighbor-net, consensus networks, super networks methods or methods for computing hybridization or simple recombination networks. It uses the NEXUS file format. The splits graph is defined using a special data block (SPLITS block). See also *Phylogenetic tree viewers *Dendroscope Dendroscope is an interactive computer software program written in Java for viewing Phylogenetic trees. This program is designed to view trees of all sizes and is very useful for creating figures. Dendroscope can be used for a variety of analyse ... * MEGAN References External links SplitsTree homepage(New Website for informations about SplitsTree)for the latest version (4.15) and manual (June 201 ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Dendroscope
Dendroscope is an interactive computer software program written in Java for viewing Phylogenetic trees. This program is designed to view trees of all sizes and is very useful for creating figures. Dendroscope can be used for a variety of analyses of molecular data sets but is particularly designed for metagenomics or analyses of uncultured environmental samples. It was developed by Daniel Huson and his colleagues at the University of Tübingen in Germany, who also created SplitsTree. See also *List of phylogenetic tree visualization software *SplitsTree *MEGAN Megan is a Welsh feminine given name, originally a diminutive form of Margaret. Margaret is from the Greek μαργαρίτης (''margarítēs''), Latin ''margarīta'', "pearl". Megan is one of the most popular Welsh-language names for women in ... References External links Dendroscope homepage hosted at the University of Washington Phylogenetics software {{science-software-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
.jpg)